Two sgRNA guide sequences specifically targeting pig Pax7 gene and application
A specific and sequence technology, applied in the field of plant genetic engineering, to achieve the effect of effective modification and mutation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
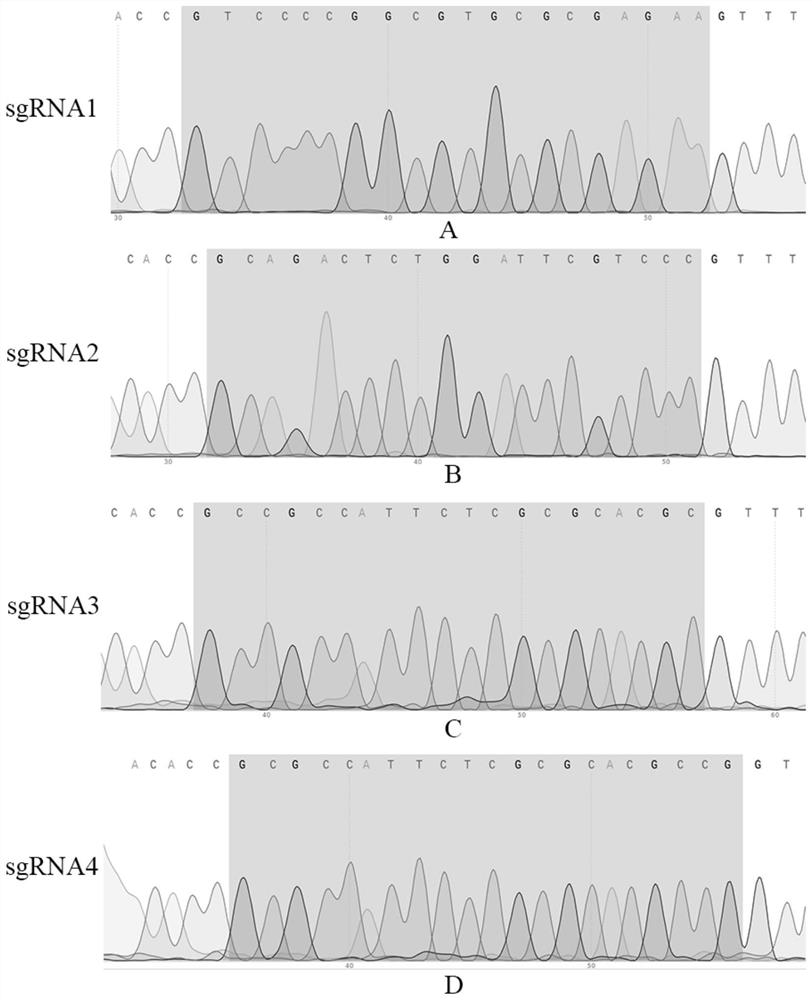
[0056] Example 1 Obtaining of sgRNA sequence
[0057] According to the genome sequence of the pig Pax7 gene (accession number AEMK02000045.1), four Pax7-sgRNAs were designed near its start codon ATG, and CACC bases were added to its 5' end to obtain the Pax7-sgRNA-F primer sequence; at the same time Obtain its corresponding DNA complementary strand according to the guide sequence, and add AAAC bases at the 5' end to obtain the Pax7-sgRNA-R primer sequence; synthesize the above-mentioned Pax7-sgRNA-F and Pax7-sgRNA-R respectively, and synthesize the two The oligonucleotides were denatured at 95°C for 10 minutes and annealed at 65°C for 50 minutes to form double-stranded DNA with cohesive ends. See Table 1 for the specific oligonucleotide sequences.
[0058] Table 1 The oligonucleotide sequences corresponding to the four sgRNAs
[0059]
Embodiment 2
[0060] Example 2 Construction of the vector expressing sgRNA
[0061] 1. Construction of linearized vector: Plasmid vector pX330 (Addgene) was digested with BbsⅠ, the digestion system was 50uL system: BbsⅠ2uL, 10X NEB Buffer 5uL, plasmid 2ug, made up to 50uL with double distilled water. Digestion conditions: enzyme digestion at 37°C for 2 hours; after agarose gel electrophoresis of the digested products, gel recovery was carried out using a gel recovery kit (Omega). See the instructions of the aforementioned kit for the recovery method.
[0062] 2. Ligation: Ligate the recovered pX330 digested with the double-stranded DNA prepared in Example 1. The ligation system is 10uL. Make up to 10uL with double distilled water; the connection condition is 16°C for 2h.
[0063] 3. Transformation: Transform the ligation product into Escherichia coli competent DH5α. The specific transformation method is: take out Escherichia coli competent DH5α at -80°C and dissolve on ice; Put it in a ne...
Embodiment 3 4
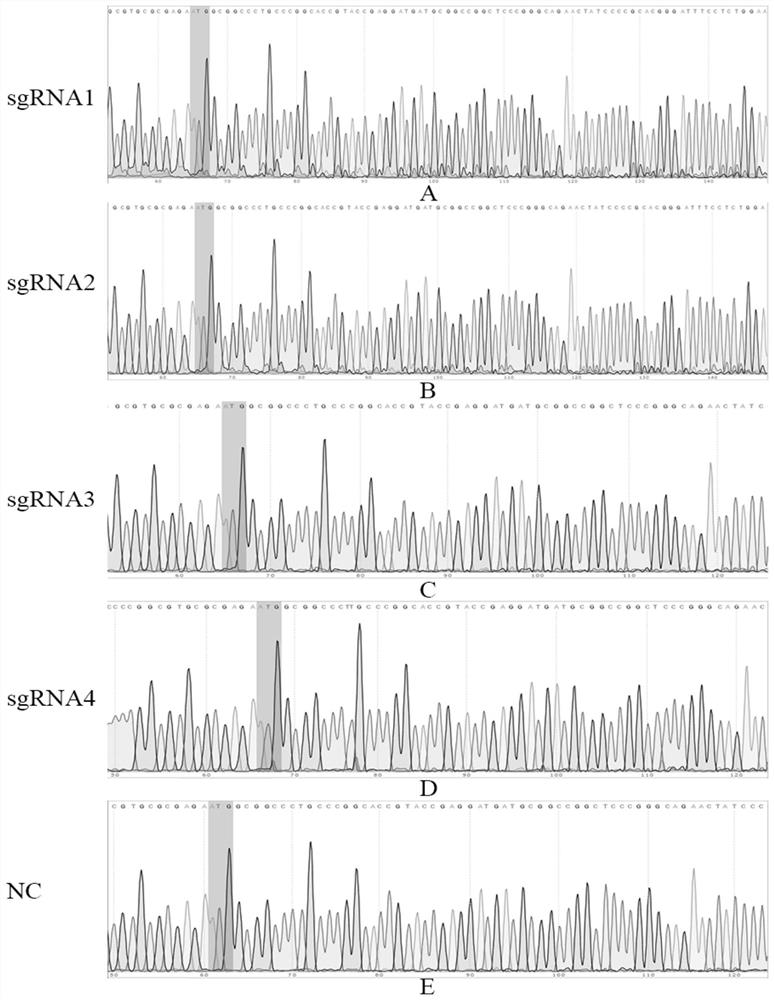
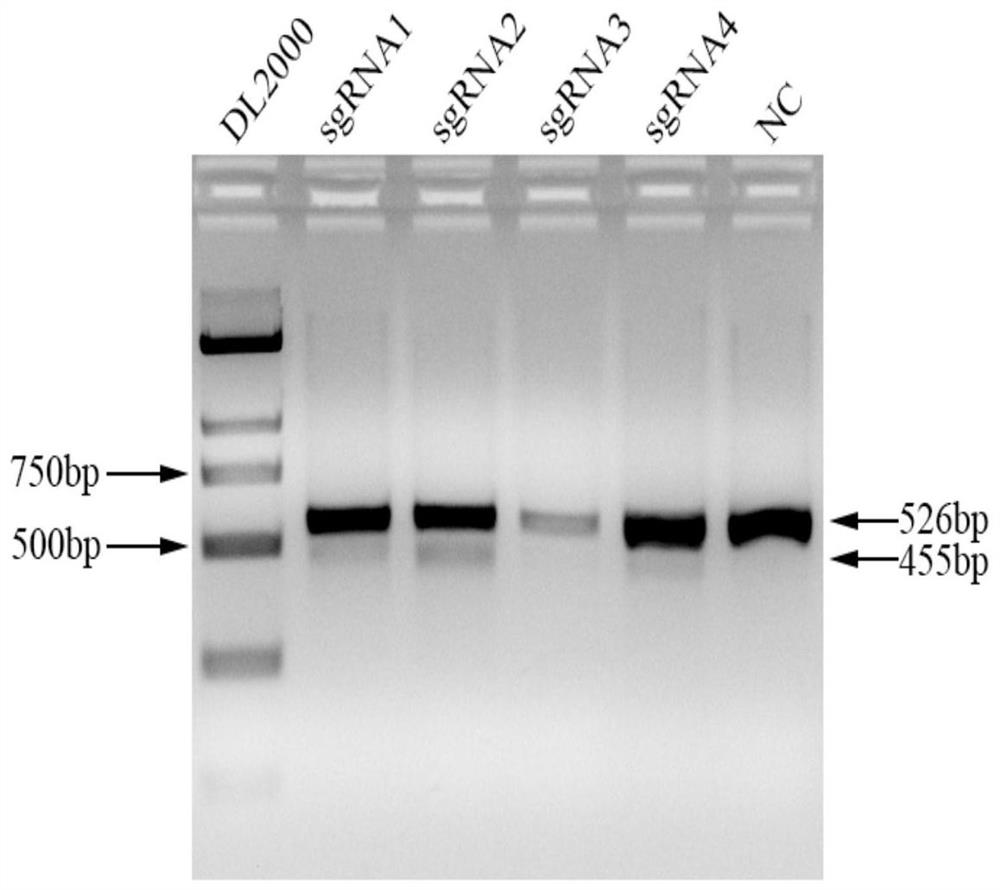
[0065] Example 3 The specific targeting verification of four sgRNAs
[0066] 1. Cell culture and collection: PK15 cells (ATCC) were revived with complete medium, and transfected after 2-3 passages. One day before transfection, 5×10 5 ~8×10 5 Cells were seeded into 6-well plates and transfected when the density reached 70%.
[0067] 2. Transfection: For specific steps of transfection, refer to the operation instructions of Lipo2000. The four vectors constructed in Example 2 were transfected with lipo2000 (Invitrogen), and the cells were collected after 48 hours, and the NC plasmid (empty plasmid) was used as a control.
[0068] 3. Screening of specific targeting sgRNA: According to the position of the designed sgRNA sequence, design identification primers, the primer sequence is shown in Table 2, extract cell DNA after 48h, and carry out PCR amplification with identification primers, the reaction system is 50uL: 10XBuffer: 5uL, dNTP: 4uL, Lataq: 0.5uL, F: 1uL, R: 1uL, DNA: 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com