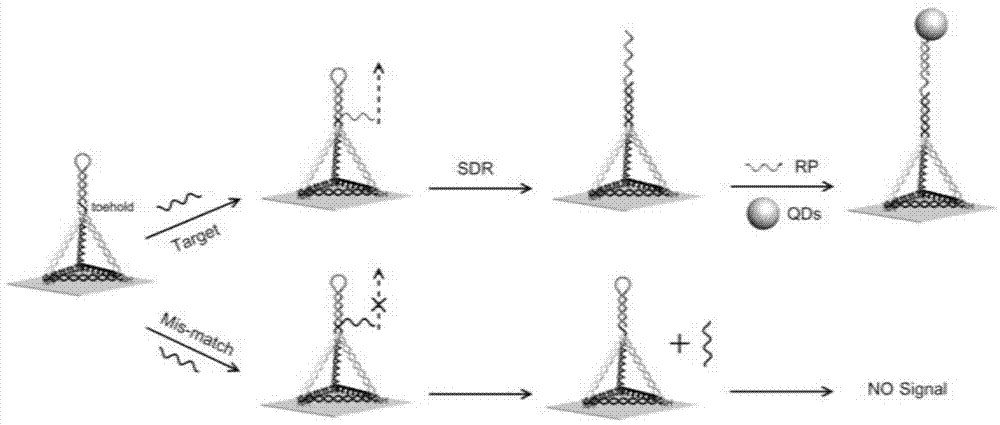
Single molecular detection method of let-7a in human lung cells
A let-7a, single-molecule detection technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as failure to achieve quantitative detection of targets and no practical application value, and achieve great application potential, The effect of good specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Single molecule detection of target let-7a
[0047] Specific steps are as follows:
[0048] (1) DNA tetrahedron assembly:
[0049] Dilute the four DNA strands (A-HP, B-NH2, C-NH2 and D-NH2) to 50 μM respectively, take 2 μL each into 92 μl TM buffer, mix well, anneal at 95 ° C for 2 min, and place in ice bath to obtain DNA tetrahedron solution, wherein the concentration of the DNA tetrahedron is 1 μM.
[0050] Electrophoresis verification: use 9.6% polyacrylamide gel electrophoresis to verify the DNA tetrahedron solution.
[0051] In order to characterize the assembly of the DNA tetrahedron, this embodiment carried out polyacrylamide gel electrophoresis verification, such as figure 2 , compared with other single-stranded structures and structures lacking one or two strands, the DNA tetrahedron moves slowly, which is consistent with previous reports and confirms the successful synthesis of the DNA tetrahedral structure. 36,37
[0052] (2) Construction of ...
Embodiment 2
[0057] Embodiment 2: the sensitivity investigation of method
[0058] Specific steps are as follows:
[0059] Same as steps (1) and (2) described in Example 1.
[0060] (3) Sensitivity investigation of miRNA detection
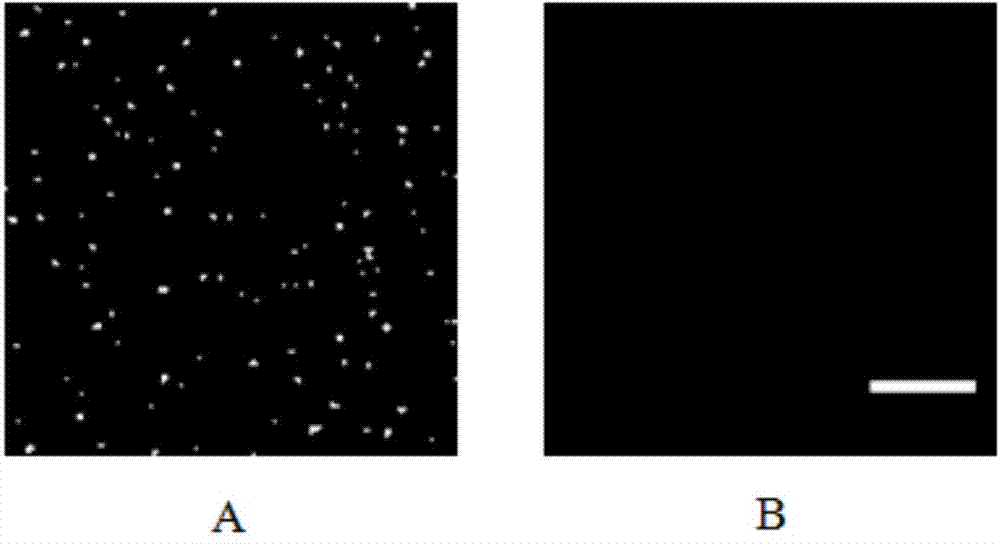
[0061]First, 50 μL of miRNA at different concentrations (1.0pM, 0.8pM, 0.5pM, 0.3pM, 0.1pM, 0.05pM and 0.005pM) were added to the prepared substrate, and incubated at 37°C for 2h. After washing with PBS-T and PBS buffer three times respectively, 50 μL of bio-RP (0.1 nM) was added and incubated at 37° C. for 2 h. After washing with PBS-T and PBS buffer three times, 50 μL of QDs (0.1 nM) was added and incubated at 37° C. for 2 h. After washing with PBS-T and PBS buffer three times, 50 μL of PBS buffer was added, imaging was performed under a fluorescence episcopic microscope, and miRNA was quantified by counting the number of fluorescent spots.
[0062] The present invention detects miRNAs of different concentrations in the buffer solution, such as Figure 4...
Embodiment 3
[0063] Embodiment 3: the specificity investigation of method
[0064] Specific steps are as follows:
[0065] Same as steps (1) and (2) described in Example 1.
[0066] (3) Specificity investigation of miRNA detection
[0067] First, 50 μL of different let-7 family member miRNAs were added to the prepared tetrahedral substrate, and incubated at 37°C for 2 hours. After washing with PBS-T and PBS buffer three times respectively, 50 μL of bio-RP (0.1 nM) was added and incubated at 37° C. for 2 h. Wash with PBS-T and PBS buffer three times respectively, add 50 μL QDs (0.1 nM), and incubate at 37° C. for 2 h. After washing with PBS-T and PBS buffer three times, 50 μL of PBS buffer was added, and imaging was performed under a fluorescence episcopic microscope. The specificity of the method was investigated by counting the number of fluorescent spots produced by different targets.
[0068] miRNAs are characterized by high sequence similarity among homogeneous groups, so it is a g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com