Primers, kits and identification methods for assisting the identification of carnivorous mite and common carnivorous mite in Malacca
A technology for auxiliary identification and kits, applied in biochemical equipment and methods, measurement/inspection of microorganisms, DNA/RNA fragments, etc., can solve the problems of time-consuming, large-scale, time-consuming and money-consuming sequences, and achieve short time-consuming , easy operation and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1, the design of specific primer pair
[0034] 1. Implementation process
[0035] Firstly, samples of common mites on grain depots and warehouses were collected, including morphologically similar carnivorous mite and common carnivorous mite. The mitochondrial cytochrome oxidase subunit I barcode (mtDNA COI barcode) sequence was obtained by genomic DNA extraction and PCR amplification, and the mtDNA COI barcode gene sequence of mites in the BOLD database and GenBank were collected. On this basis, the DNAMAN software was used to compare and analyze the mtDNA COI barcodes of different species, and to find the specific sequence segments of the Malacca carnivorous mite and the common carnivorous mite. The specific primer pairs of Melaka carnivorous mite and common carnivorous mite were artificially designed, and the primers were evaluated and checked by Oligo software and Blast program. Under the standard PCR reaction system and reaction conditions, the specific...
Embodiment 2
[0056] Composition and use of embodiment 2 kit
[0057] The kit includes the primer pair (Cma-F / Cma-R) designed in Example 1 for identifying carnivorous mite in Malacca and the primer pair (Cer-F / Cer-R) for identifying common carnivorous mite, 10×PCR Buffer (with Mg 2+ ), dNTPs, Taq DNA polymerase and DEPC water, the above reagents were purchased from Tiangen Biochemical Technology (Beijing) Co., Ltd.
[0058] Use of the kit:
[0059] (1) Extract the genomic DNA of the mite to be tested, which can be extracted using commercial kits or methods known in the prior art;
[0060] (2) Using the genomic DNA extracted in step (1) as a template, use the above two pairs of primers to carry out PCR amplification reaction respectively; reaction system (25μL): Taq enzyme (5U / μL) 0.2μL, 10×PCR Buffer (containing Mg 2+ ) 2.5 μL, dNTPs (2.5 μM) 2 μL, upstream and downstream primers 0.5 μL each (one PCR with primer pair Cma-F / Cma-R, another PCR with primer pair Cer-F / Cer-R), template 2 μL, ...
Embodiment 3
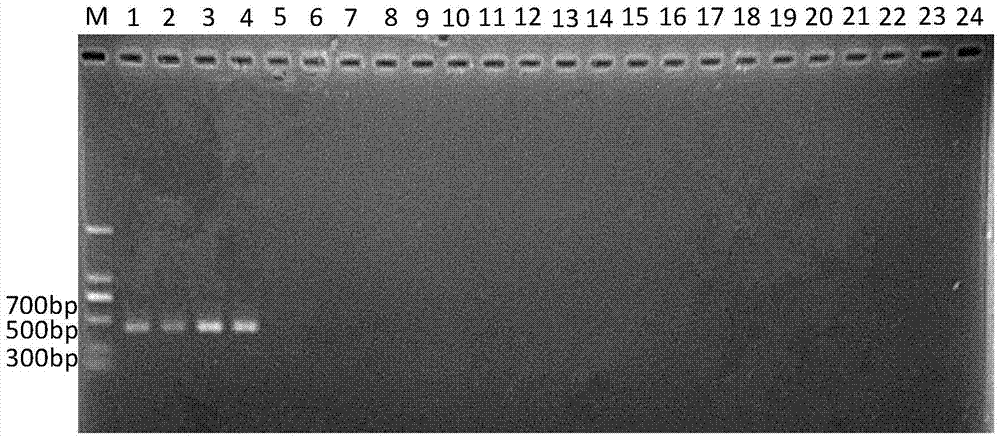
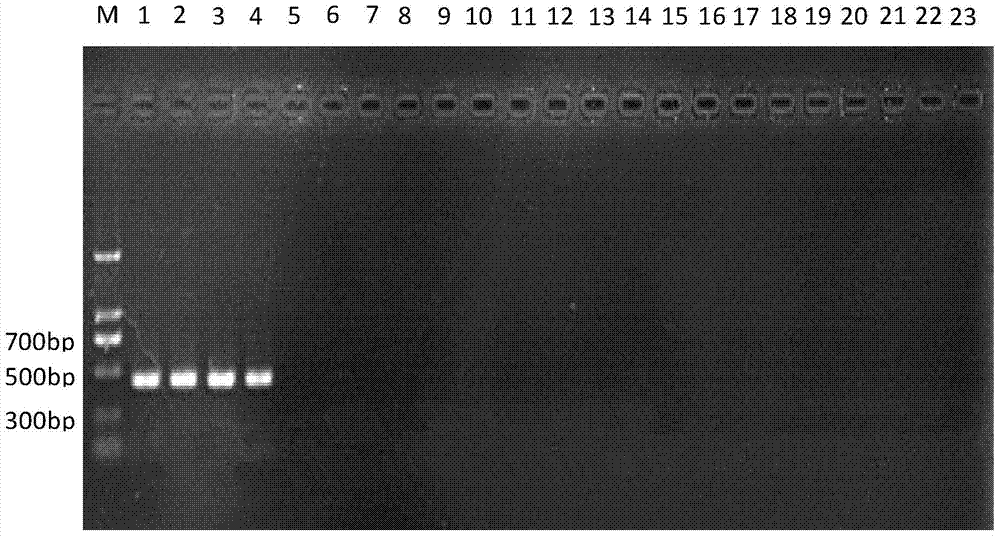
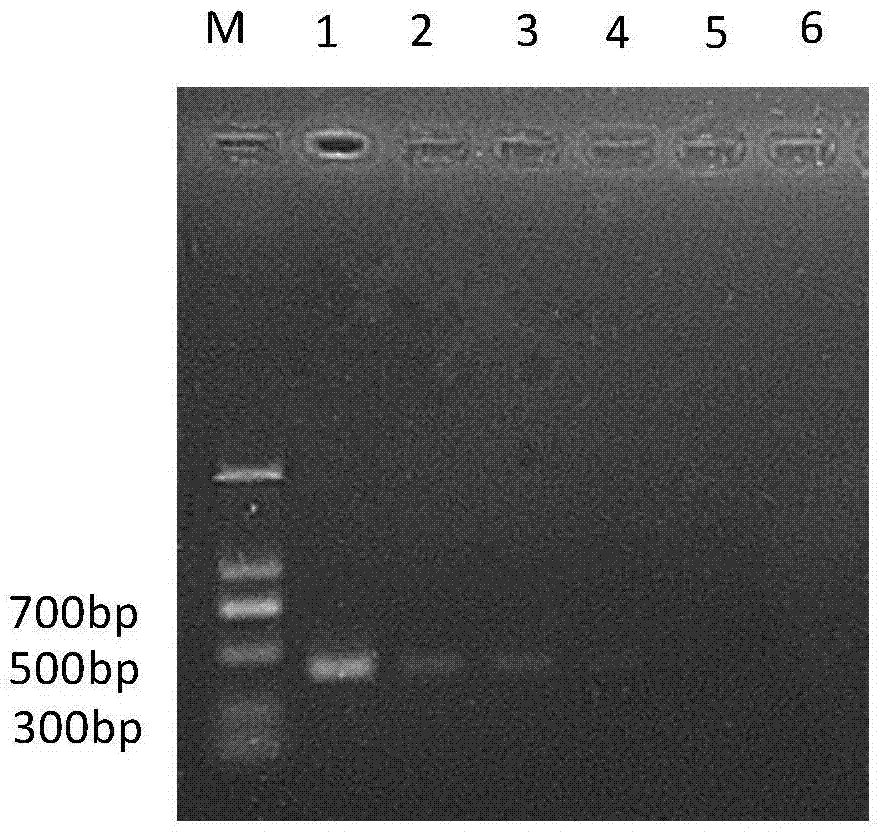
[0063] Embodiment 3, specificity experiment
[0064] Cheyletus eruditus, Schrank, Cheyletus malaccensis Oudemans, Acarus siro L., Tyrophagus putrescentiae (Schrank), Tyroborus lini Oudemans, Aleuroglyphus ovatus (Troupeau), Lepidoglyphus destructor (Schrank), Glycyphagus domesticus (De Geer), Chortoglyphus arcuatus (Troupeau), and Carpoglyphus lactis (L.): preserved in the Scientific Research Institute of the National Grain Administration.
[0065] 1. Experimental samples
[0066] The samples of 10 kinds of storage mites were collected from my country and the Czech grain depots, among which the Malacca carnivorous mites were from the Haikou grain depot in Hainan, my country, and the rest of the mite samples were from the Czech Republic. The samples were collected in 2012 and stored at -80°C in In absolute ethanol, the common carnivorous mite and the malacca carnivorous mite were live insects raised in the laboratory, and the specific information is shown in Table 2.
[0067] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com