Molecular diagnostic test for cancer
A technique for cancer, testing samples, applied in the field of molecular diagnostic testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0356] Example 1 : Tissue processing, hierarchical clustering, subtype identification and classifier development
[0357] tumor material
[0358] Identification of exemplary expression signatures from gene expression analysis of a cohort of grossly dissected epithelial serous ovarian tumor FFPE tissue samples from NHS Lothian and University of Edinburgh.
[0359] The protocol for histological classification of epithelial ovarian cancer to define serous, endometrioid, clear cell, and mucinoid histology was recently updated. One consequence of this progression is that many tumors previously classified as endometrioid are now classified as serous. (McCluggage, W.G. "Morphological subtypes of ovarian cancer: a review with the mphasison new developments and pathogenesis," Pathology 2011 Aug; 43(5):420-32). The serous samples used in this study belonged to a larger collection of epithelial ovarian cancer samples with full histology collected between 1984 and 2006. Pathology t...
Embodiment 2
[0410] Example 2 : In silico validation of angiogenic subtypes and angiogenic classifier models
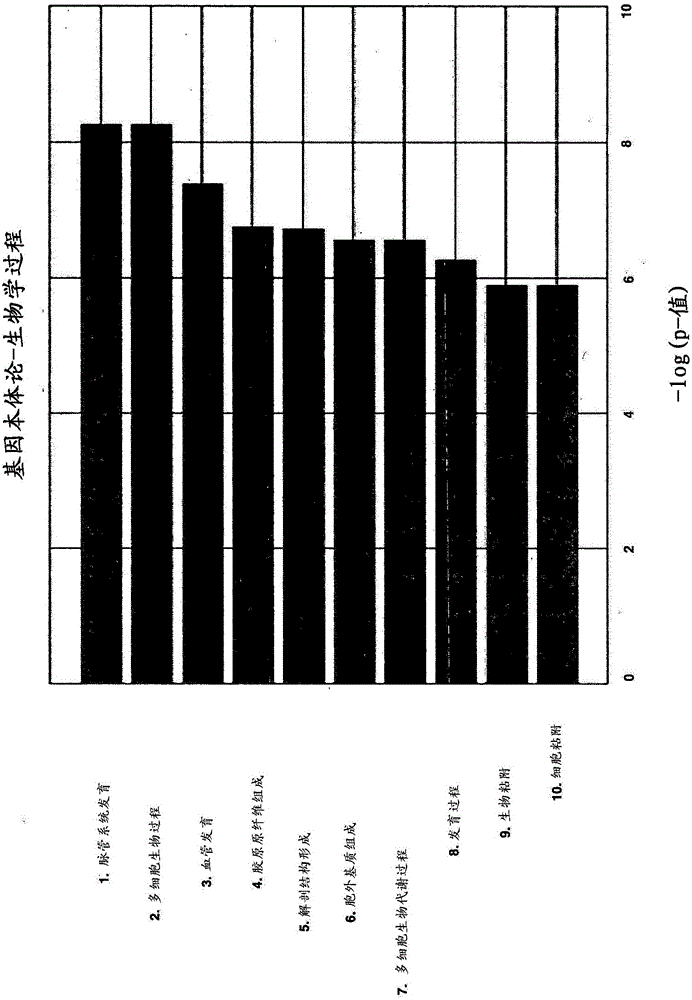
[0411]The 25-gene (initial protocol) angiogenesis classifier model and the 45-gene (reclassified scheme) performance of an angiogenesis classifier model. AUC is a statistic calculated based on observed disease scores and is a measure of the power of using a classifier model to predict a phenotype (Wray et al., PLoS Genetics Vol. 6, 1-9). AUC0.5 is common for random classifiers, and AUC1.0 would represent perfect class separation. Therefore, to determine whether an angiogenic subtype classifier model could predict patients who would respond to and be selected for an antiangiogenic ovarian cancer drug class as monotherapy or in combination with standard of care therapy, the following assumptions were made: The AUC after internal application of these data sets should be higher than 0.5, and the lowest confidence interval is higher than 0.5.
[0412] Classifier Models Applied...
Embodiment 3
[0414] Example 3 : Identification and in silico validation of the "non-angiogenic" subgroup of ovarian cancer
[0415] Classification of tumors into 'non-angiogenesis' sample groups
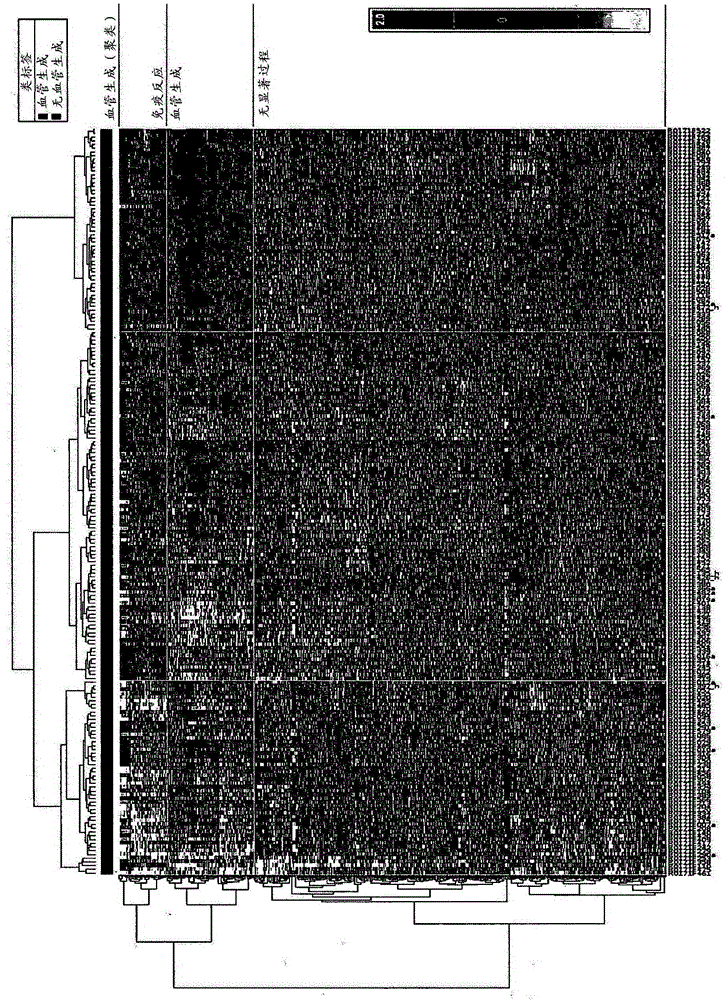
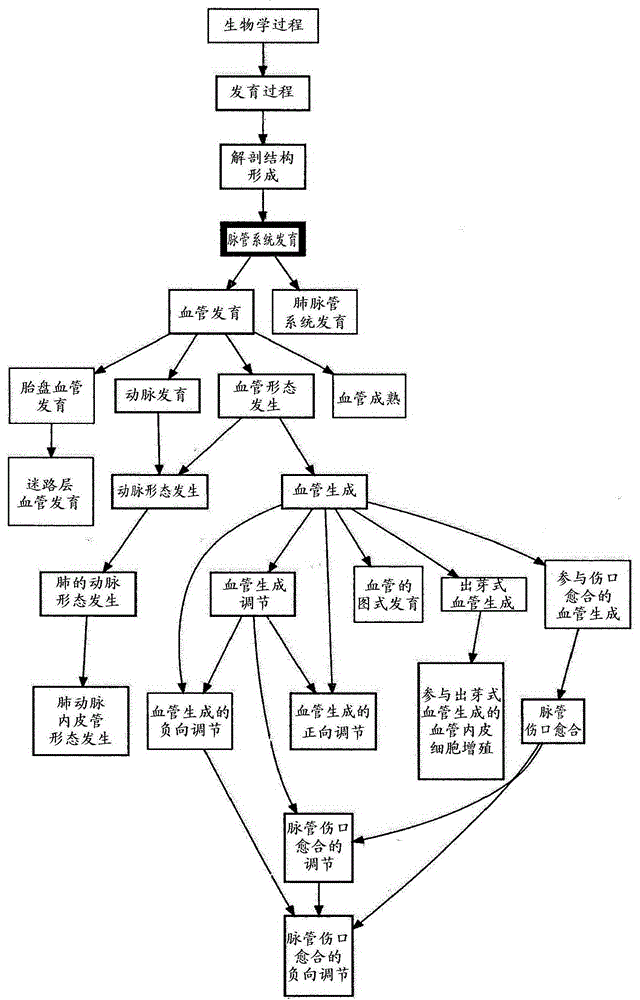
[0416] The expression of angiogenic genes in probeset cluster 2 was downregulated in all samples in sample cluster 1 of the hierarchical clustering of 265 samples newly classified as serous ( Image 6 , the blue scale marked at the bottom). from such as Figure 10 Of the samples from cluster 1 and cluster 3 pooled together as shown in , these samples in cluster 2 had a better prognosis than the remaining serous samples. This suggests that this group is defined by the downregulation of expression of the angiogenic genes identified in Table 1B. Patients with down-regulated genes involved in angiogenesis and thus this subgroup were referred to as the "non-angiogenesis" group. This phenotype has also been identified in ER+ and ER- breast cancers, as can be found in Figure 11A The intermedia...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com