Assay and method for predicting therapeutic efficacy of immunoglobulin therapy in individual patients with relapsing remitting multiple sclerosis (RR-MS)
An immunoglobulin and individual technology, applied in the direction of biochemical equipment and methods, microbial determination/testing, etc., can solve problems such as unclear underlying mechanisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
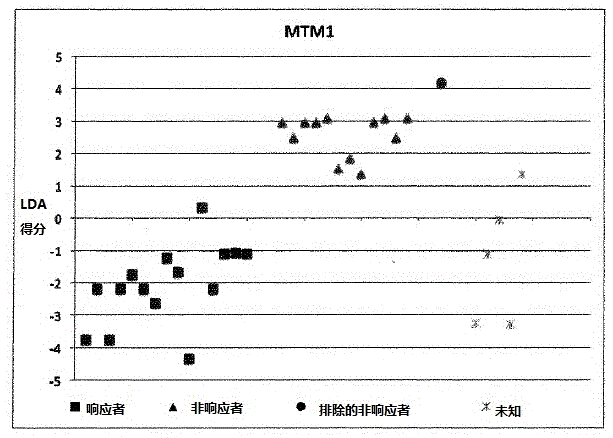
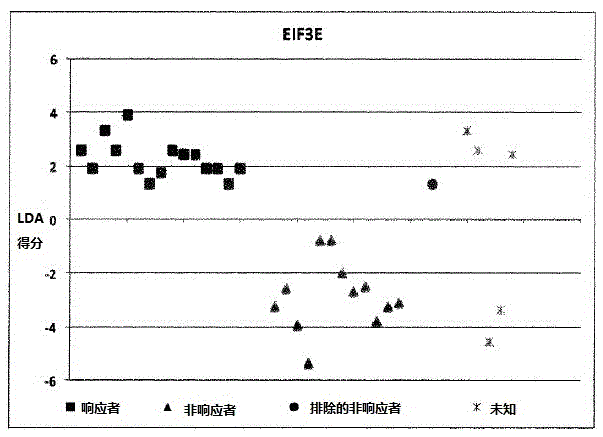
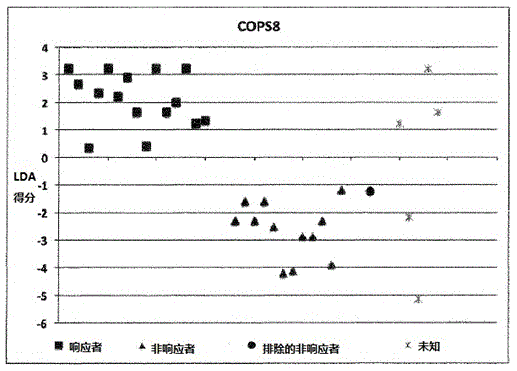
[0175] Example 1: Calculation of LDA scores involving 9 individuals:
[0176] ●Determination of SNP results
[0177] The homozygous SNP combination of interest associated with the KLHDC8A gene is represented by dbSNPRSID rs7549293-rs10751436-rs913723-rs913722 with the SNP combination (in the same order as the given dbSNPRSID) CC-TT-AA-TT, which indicates non-responders and Contribute "1" to individual LDA score calculation;
[0178] The homozygous SNP combination of interest associated with the PRDM9 and OR9Q1 genes is represented by dbSNPRSIDrs13182871-rs7701403-rs12576939-rs1376486 with the SNP combination GG-AA-TT-GG (in the same order as the given dbSNPRSID), which indicates a non-response and contribute "1" to the individual LDA score calculation;
[0179] The homozygous SNP combination of interest associated with the ADAMTS9 gene consists of dbSNPRSID rs9820942-rs6780659-rs6445415-rs11721258 with the SNP combination GG-CC-AA-TT-AA-GG-TT-AA (in the same order as the g...
Embodiment 2
[0202] Calculations involving 2-8 LDA scores were performed in the same manner as in Example 1.
[0203] This method makes it possible to identify the responsiveness / non-responsiveness of a person to any immunoglobulin product suitable for in vivo use in diseases that can in principle be treated by immunoglobulins, such as intravenous, Those immunoglobulin products for subcutaneous, intramuscular, intraocular, intrathecal, buccal, topical or inhalation administration of diseases such as immune-mediated inflammatory diseases, autoimmune diseases, allergies, graft-versus-host reactions and prophylaxis Transplant rejection, multiple sclerosis of any kind or any other demyelinating neurological disease, or relapsing-remitting multiple sclerosis.
[0204] The method also allows the prediction of the MS patient's likelihood of relapse and / or the rate of progression of the disease in the patient's disability and / or function (measured by a clinical scale such as, but not limited to, t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com