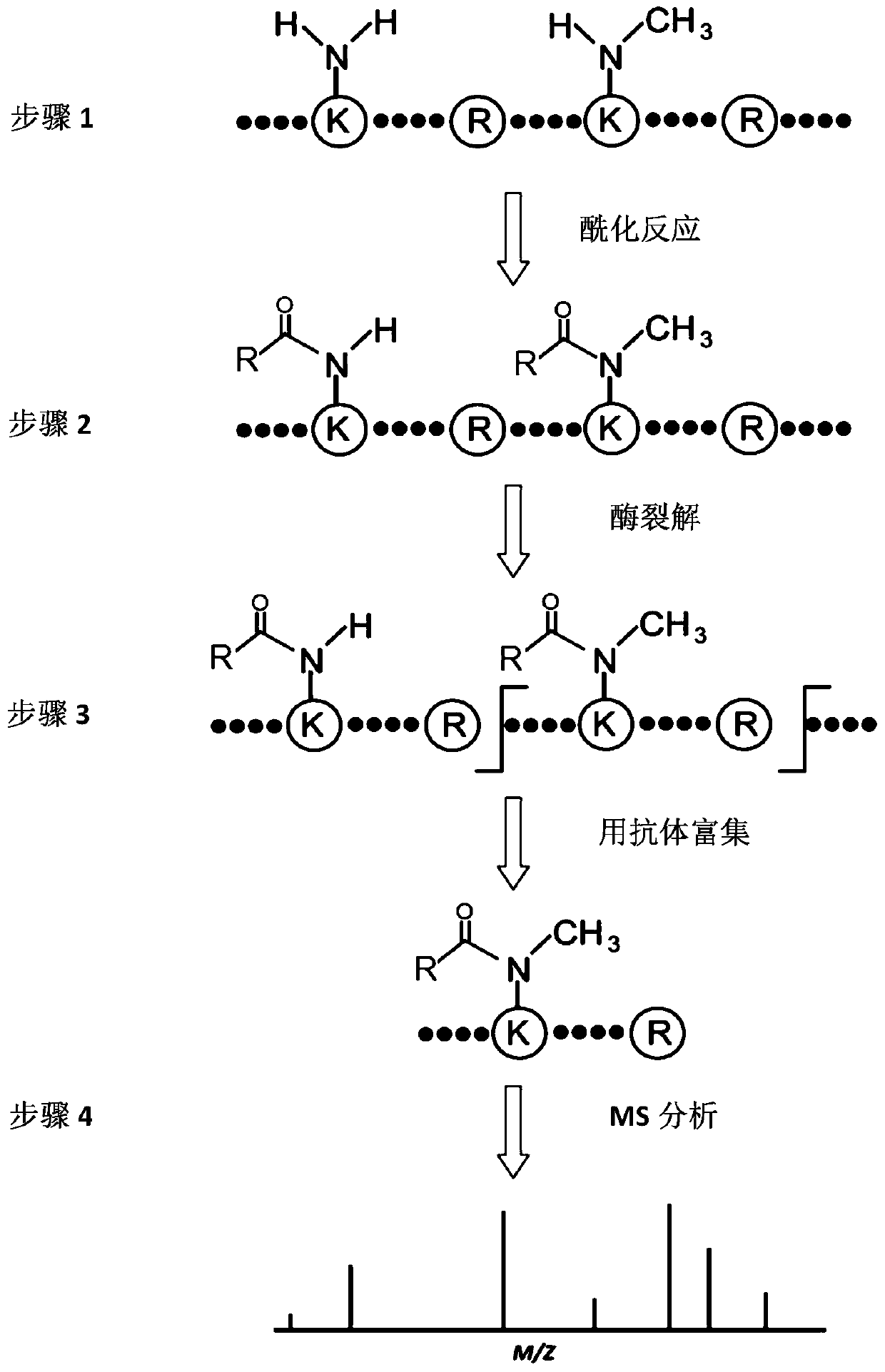
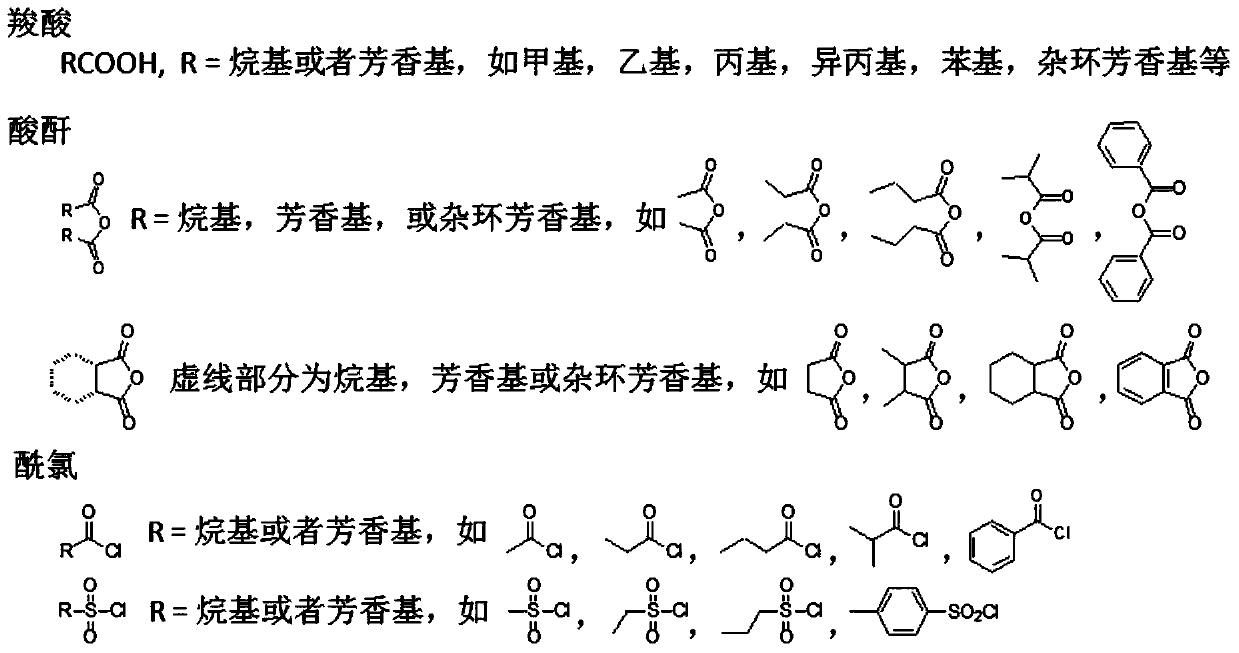
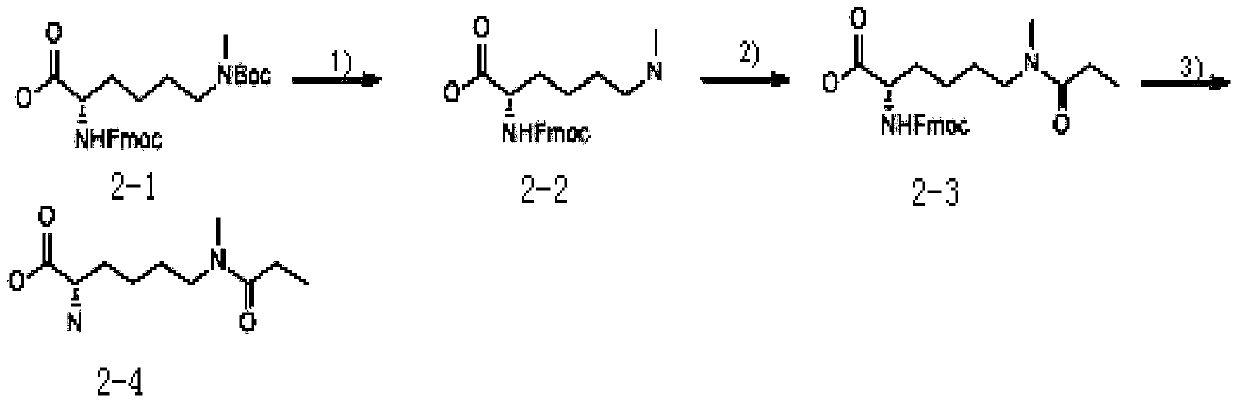
Method for identifying single methylated modification of lysine epsilon-amino group side chain
A lysine residue, lysine technology, applied in the field of detection and diagnosis, can solve problems such as antibody difficulties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0059] The present invention uses a specific embodiment to identify or quantify the presence of lysine monomethyl-modified peptides or proteins in cells or tissues, and this enumeration is only exemplary to illustrate how the method of the present invention is implemented, and The present invention cannot be limited in any way. Those skilled in the art can make any improvements and changes to the present invention without departing from the essence of the present invention, but such changes are all included in the scope of the claims of the present invention.
[0060] Example 1: Stable isotope labeling of methionine, in vitro propionylation derivatization and proteolysis
[0061] 1.1 Stable isotope labeling of methionine
[0062] The cells used in the present invention include HeLa cells (cervical cancer cell line), K562 cells (chronic myeloid leukemia cells), SW620 cells (colon cancer cells), A549 cells (lung cancer cells) and SMM7721 cells (hepatoma cells), cultured in R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com