Method for identifying prunus persica plant species based on SSR molecular markers
A molecular marker, almond technology, applied in the molecular marker field of molecular biology, can solve problems such as difficulty in classification, cultivation and variety selection of almonds, homonymic or synonymous, unclear variety pedigree, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
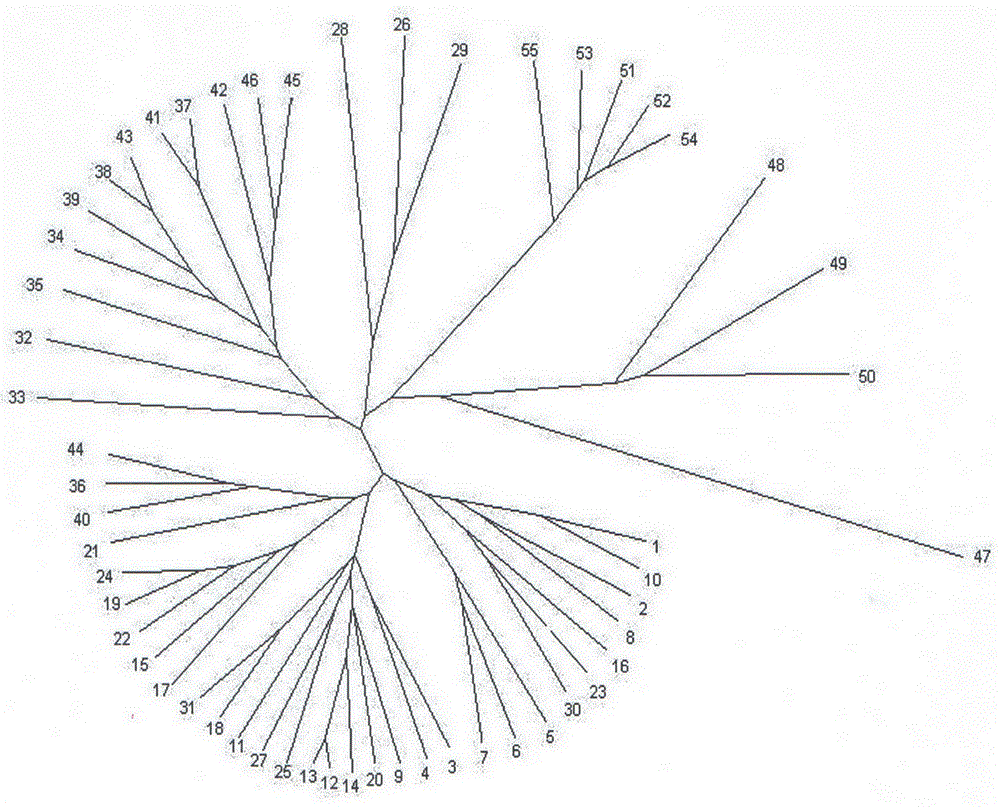
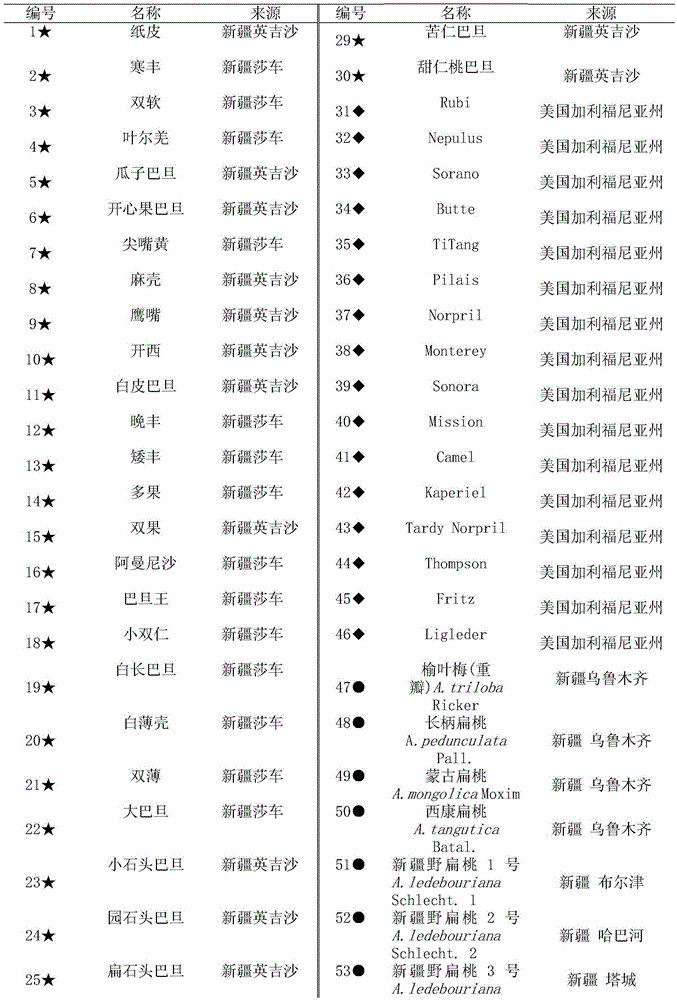
[0042] Preparation of almond germplasm materials: 5 Xinjiang wild almonds collected from different distribution populations in Tacheng and Altay, Xinjiang, China (in 5 different distribution centers of Burqin, Habahe, Tacheng, Tuoli, Randomly selected in the area, respectively numbered as Xinjiang Wild Almond No. 1, No. 2, No. 3, No. 4, and No. 5 individual plants); collected from Xinjiang local cultivated almond varieties in Yingjisha and Shache County, Kashgar region, southern Xinjiang, China 30 parts; 16 parts of excellent cultivated almond varieties imported from California, USA, collected from Shache County, Kashi District, southern Xinjiang, China; Mongolian almonds, Xikang almonds, long-stalked almonds and Yuyemei collected from the Forest Fruit Resource Garden of Xinjiang Academy of Forestry Sciences 4 copies of the germplasm resources of almonds; a total of 55 sample materials, which basically cover the germplasm resources of almonds. The samples were frozen in liquid...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com