Nucleic acid inducing RNA interference modified for preventing off-target, and use thereof
一种RNA干扰、脱靶效应的技术,应用在用于阻止脱靶效应而变形的RNA干扰诱导核酸及其用途领域,能够解决减少命中目标抑制效率等问题,达到缓解不准确性及副作用、防止脱靶效应的效果
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Comparison of target gene expression inhibition effect and off-target effect of siRNA molecules replaced by deoxyribonucleotide spacer (dSpacer)
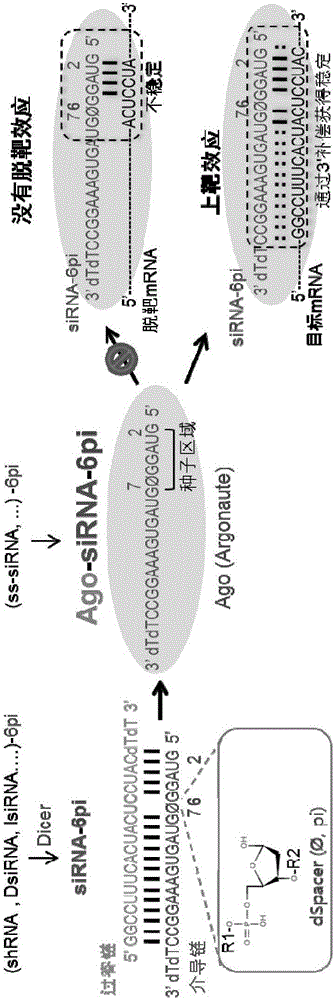
[0069] Such as figure 1 For the example shown in the diagram, the inventors speculate that applying a base-pairing-incapable spacer deformation to the 5' seed region of all RNAi-inducing nucleic acids bound to the AGO protein could completely prevent off-target effects while improving target gene repression effect (on-target effect), the inventors' model based on the microRNA recognition mechanism, the transitional nuclear model (NatStructMolBiol.2012Feb12; 19(3):321-7), focuses on the 5' seed region, especially the so-called The present RNA interference-inducing nucleic acid is invented by base pairing at the 6th nucleotide position from the 5' end of the "pivot".
[0070] First, in order to compare the on-target and off-target effects mediated by the deformed siRNA molecules according to the present invention and conventio...
Embodiment 2
[0079] Comparison of target gene inhibition and off-target effects caused by siRNA molecule ribonucleotide spacer (rSpacer) replacement
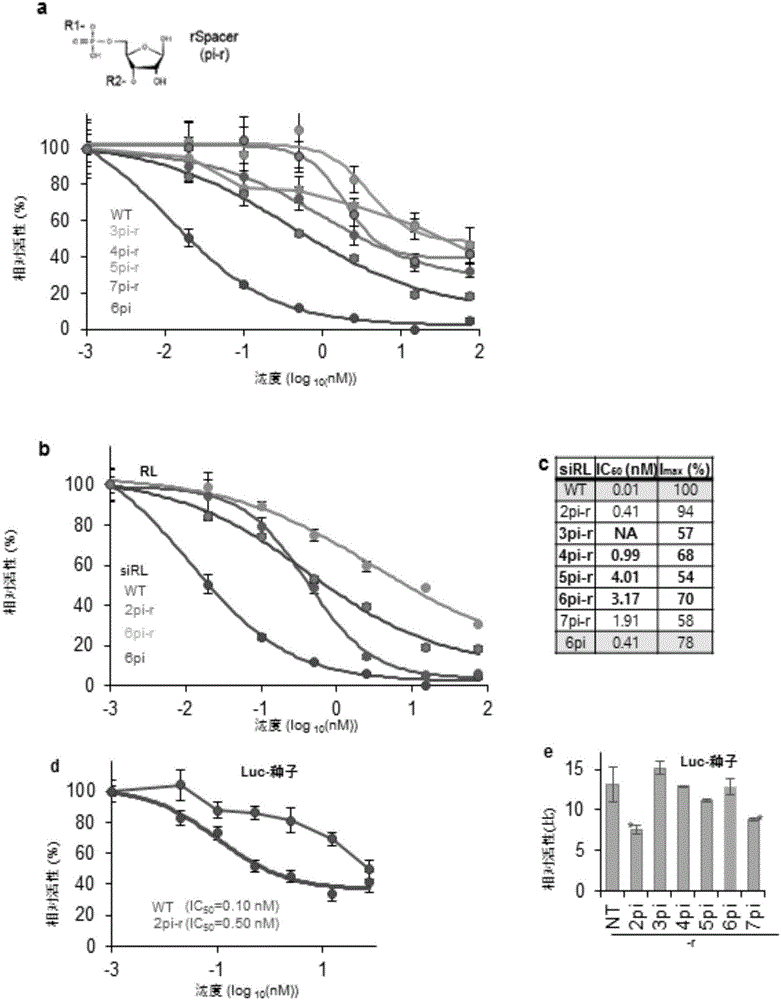
[0080] To see whether the results in Example 1 above can be replicated by a similar spacer containing a ribonucleotide backbone, the nucleotides at positions 2-7 of siRL are replaced by a ribonucleic acid spacer (rSpacer) (pi-r ), this position was previously shown to eliminate off-target effects, and here the targeting activity of siRL was detected by calculating IC50 according to the same method as in Example 1.
[0081] results, such as image 3 As shown in a and 3c, each rSpacer substitution at positions 2-7 from the 5' end showed a maximum inhibition rate (Imax) greater than or equal to 54% relative to untransformed (WT). Of these, each variant except 2pi-r (applied to site 2) had less on-target inhibitory activity than 6pi. Especially 2pi-r showed superior on-target inhibition than 6pi, but still as image 3 The inhibition of off-ta...
Embodiment 3
[0084] Comparison of target gene inhibition and off-target effects caused by implantation of siRNA molecule deoxynucleotide spacer (dSpacer)
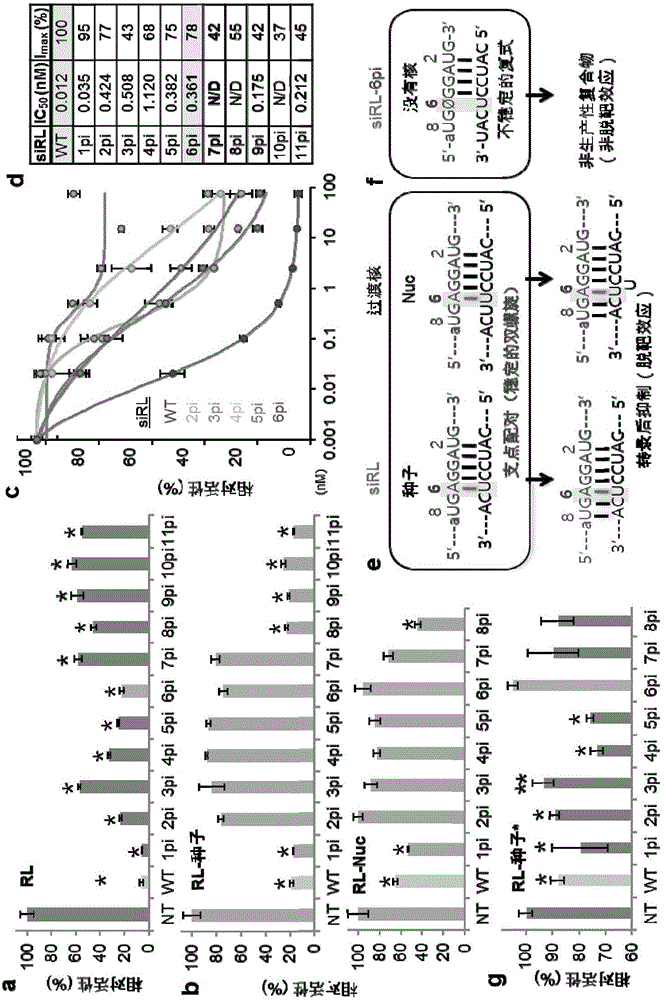
[0085] The sequence configuration in the seed region contains a deoxynucleotide spacer (dSpacer). When interacting with the target gene RNA, inconsistent matching should occur in a non-base part of the siRNA, which is passed in Example 1 and Example 2. The substitution in is made. Therefore, when a dSpacer is implanted in the seed region of siRNA, the non-base can form a bulge, such as Figure 4 as shown in a. Therefore, changes in on-target and off-target effects were investigated for the case of such dSpacer implants. Initially, the targeting effect of a siRL comprising a dSpacer implanted between positions 2-7 from the 5' end, where positions 2 to 7 are in the above example, can be detected by measuring the IC50 as in Example 1 Reported to eliminate off-targets.
[0086] Therefore, if Figure 4 In b, each implantation of dSpacer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com