CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9-system-based Saccharomyces cerevisiae genome concurrent multiplex edition vector and application thereof
A Saccharomyces cerevisiae and genome technology, applied in the field of Saccharomyces cerevisiae genome parallel multiple editing vectors, can solve the problems of long experimental cycle, time-consuming, cumbersome editing system construction, etc., and achieve the effect of simple operation and simple multi-gene editing work
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
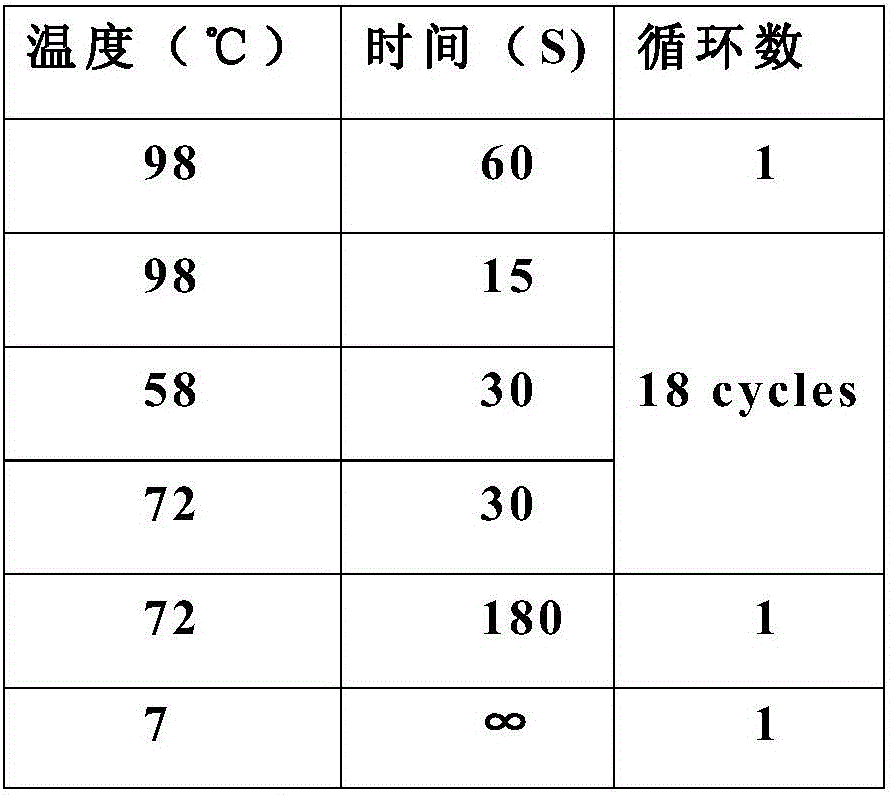
[0058] 1. Design the following sequence based on the known sequence:
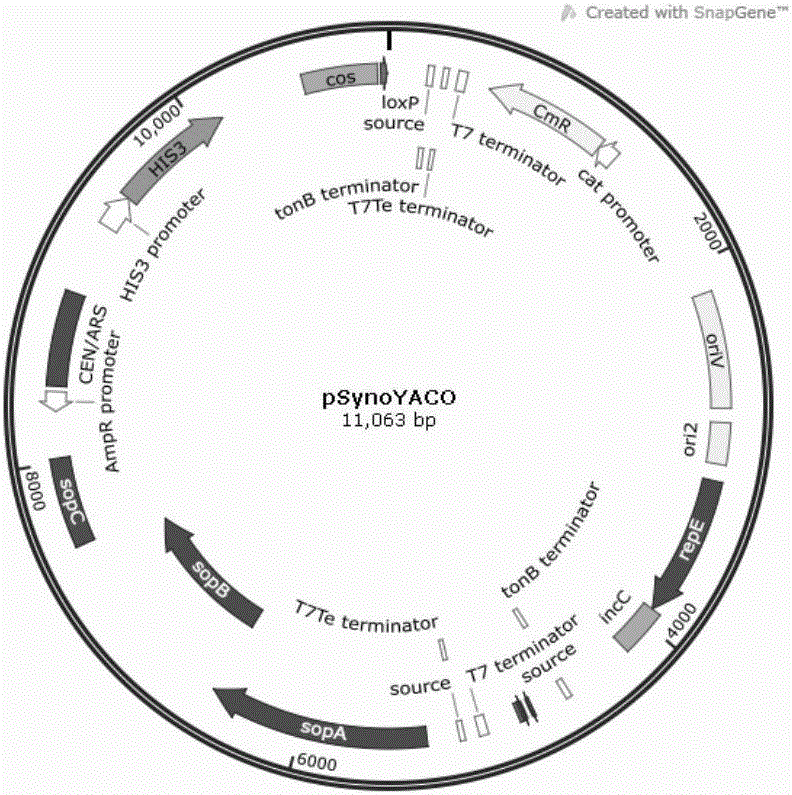
[0059] 1) Yeast Cas9 protein expression cassette (TEF1promoter-Cas9-CYC1terminitor), including TEF1 promoter, Cas9 protein, CYC1 terminator, the sequence is shown in SEQ ID NO.1, wherein: CGAACGCCATCGACTTACCAGTATGCTACTTACTAT and CAGCAGGAGCTGGACTCTACTGATGTCTGGACAGC at the beginning and end of the sequence of SEQ ID NO.1 are Cas9 protein expression cassette and pSynoYACO vector homology arm sequence; ggcgcgcc and ggcgcgcc are AscI restriction site sequences, GTTTAAAC is PmeI restriction site.
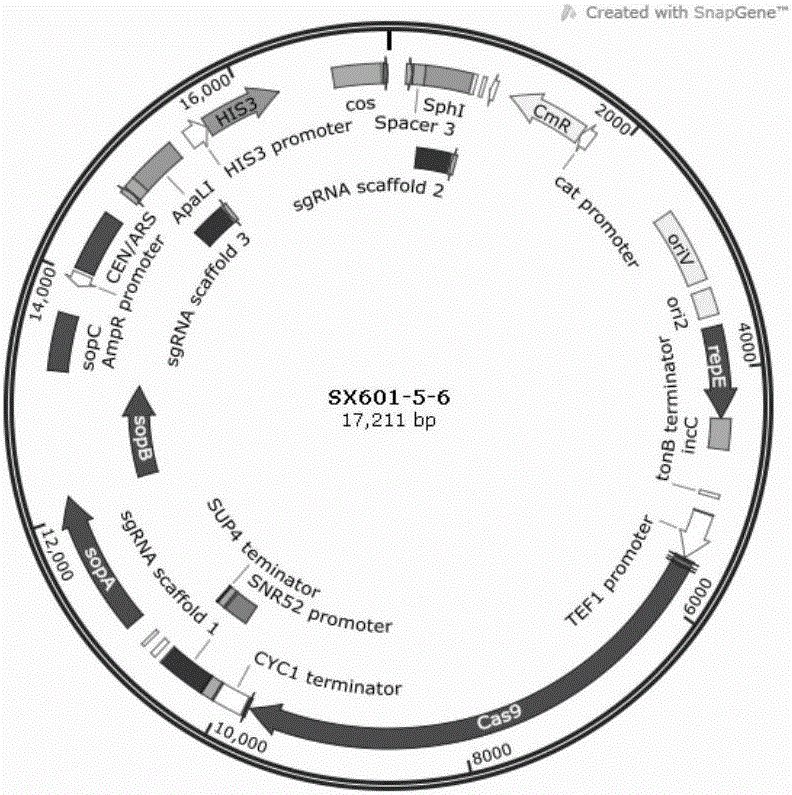
[0060] 2) The sequence of sgRNA scaffold expression box 1 (SNR52 promoter+sgRNA Scaffold+SUP4terminitor) is shown in SEQ ID NO.2, wherein: GGACGCTCGAAGGCTTTAATTTGC and gctgctaacaaagcccgaaag at the beginning and end of the sequence of SEQ ID NO.2 are sgRNA Scaffold expression box 1 and pSynoYACO-Cas9 vector The source arm sequence, GTTTAAAC is the sequence of the restriction site of PmeI.
[0061] 3) The sequence of sg...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com