Method for knocking down fish gene expression based on heat induced miR30-ShRNA
A gene expression and fish technology, applied in the field of gene expression knockdown in fish based on heat-induced miR30-shRNA, can solve the problems of poor, irreproducible results, siRNA or shRNA knockdown research technology, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0047] The zebrafish Apex1 gene information is retrieved from the database, and the ORF sequence of the zebrafish Apex1 gene is shown as SEQ ID NO.1.
[0048] Design of efficient siRNA targets against the Apex1 gene. The effective siRNA target sequence targeting the Apex1 gene contains the sequence shown in SEQ ID NO.2.
[0049] Specifically, the sequence shown in SEQ ID NO.2 is: GGAGAGGCTGACAATGGAA.
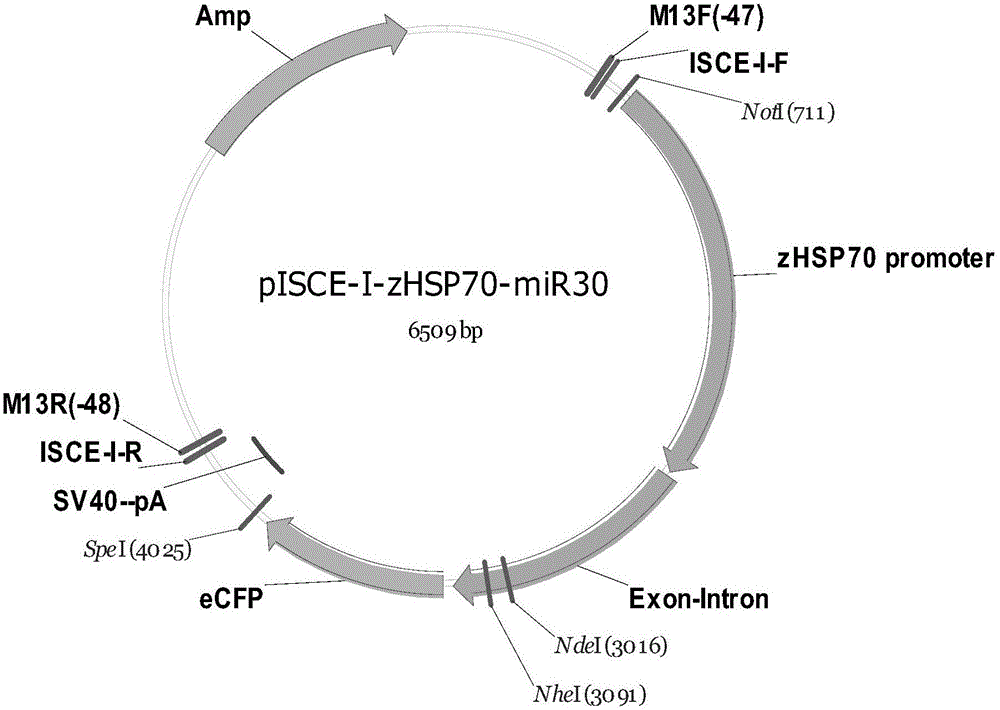
[0050] In this example, a sequence vector (pISceI-HSP70-miR30-Apex1-eGFP) that uses the heat shock promoter HSP70 to regulate the expression of downstream genes was constructed by using the miR30 interference sequence.
[0051] First, a single-stranded partial miR30 interference sequence and a single-stranded shApex1 were synthesized in vitro, and the target fragment miR30-Apex1 was inserted by annealing, with BbsI restriction sites at both ends of the target fragment. The insertion target fragment miR30-Apex1 contains the sequence shown in SEQ ID NO.3. The sequence shown in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com