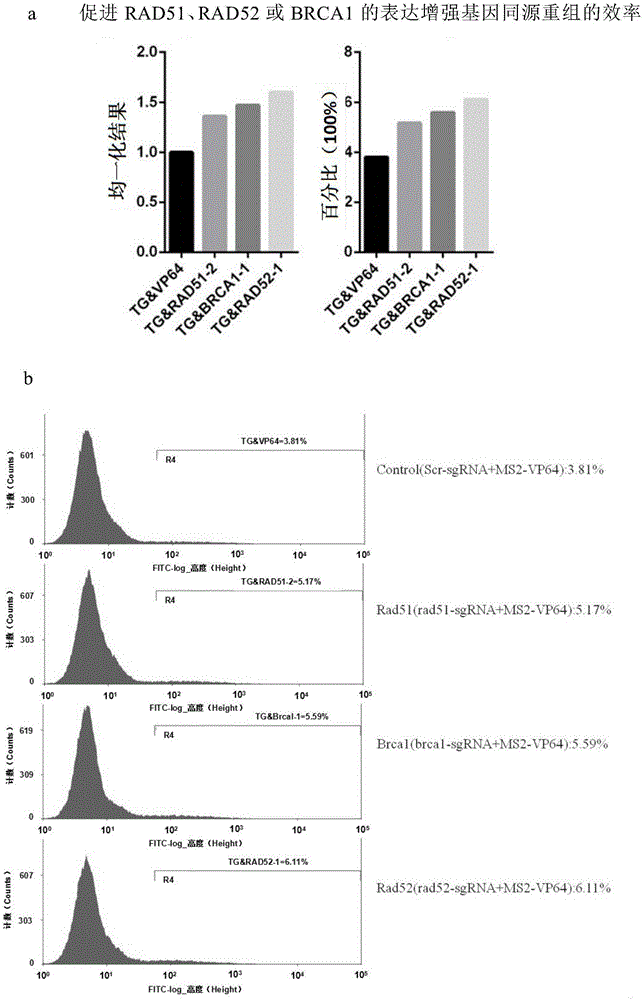
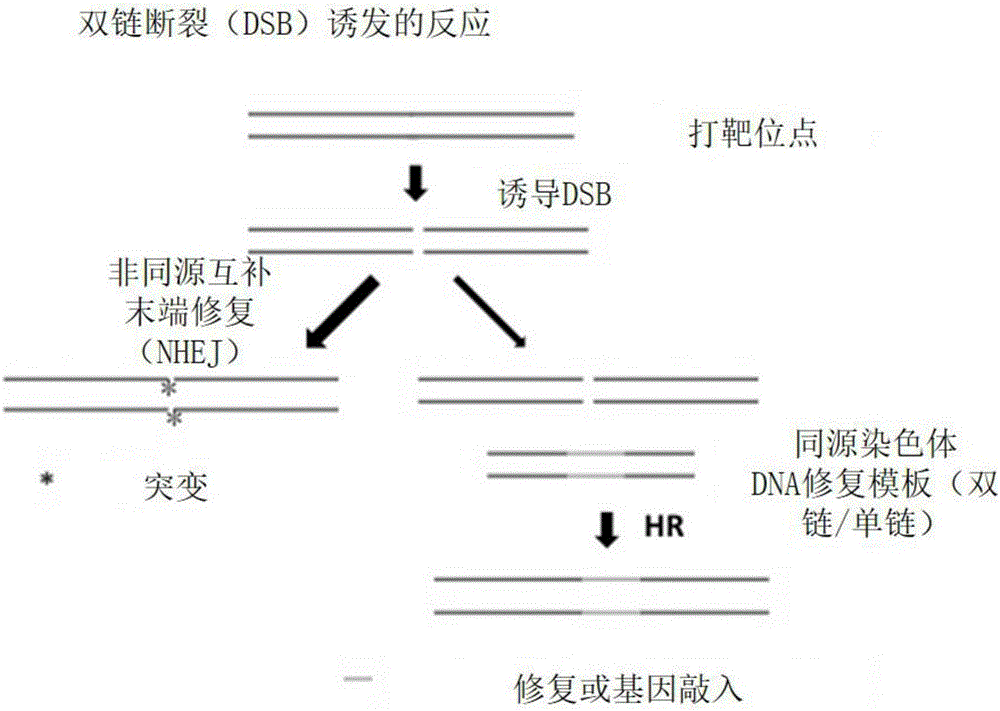
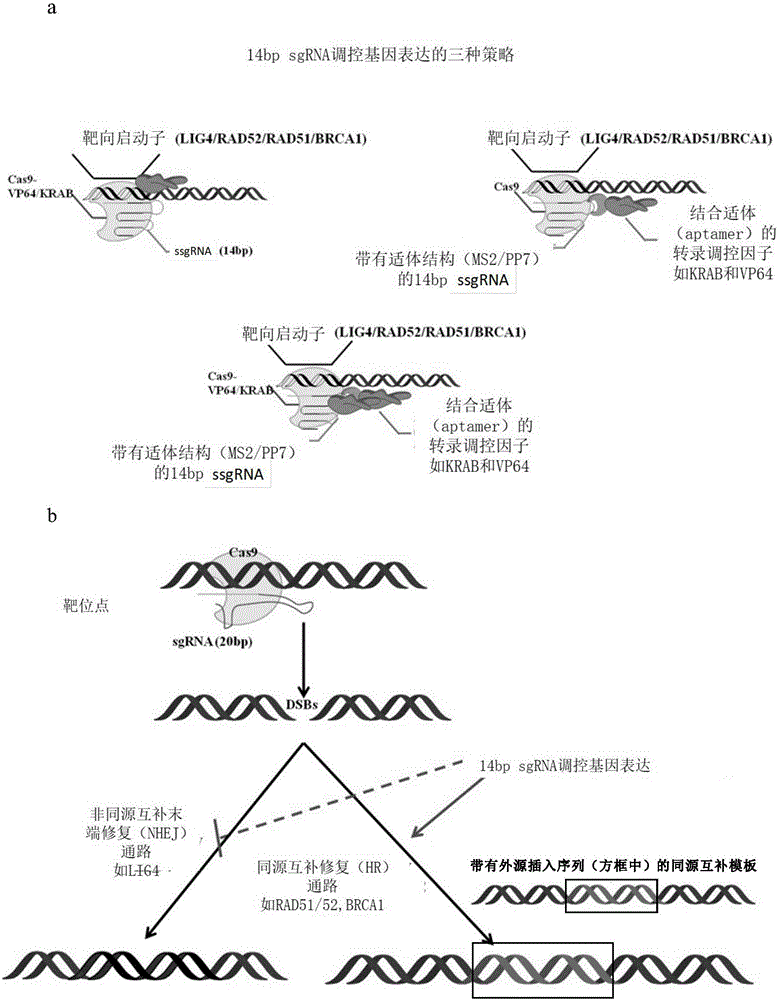
Method for gene point mutation restoration
A gene editing, target gene technology, applied in genetic engineering, biochemical equipment and methods, nucleic acid vectors, etc., can solve problems such as inability to cut, achieve the effect of improving efficiency, fine gene editing, and avoiding genome instability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1 GFP reporter gene transformation, restore the expression of GFP green fluorescent protein
[0055] 1) Transform ssgRNA to obtain ssgRNA with specific RNA hairpin structure
[0056] A unique RNA hairpin such as MS2 / PP7 is inserted at a specific site of ssgRNA (SpCas9 and SaCas9), and this type of hairpin structure can be specifically recognized by MS2 binding protein / PCP protein.
[0057] The full sequence of ssgRNA with PP7 hairpin structure is:
[0058] ccggtgagaccgagagagggtctcagttttagagctagcgagggagcagacgatatggcgtcgctccctcgttagcaagttaaaataaggctagtccgttatcaacttgcgaggggagcagacgatatggcgtcgctccctcgtaagtggcaccgagtcggtgctttttgaattc
[0059] The full sequence of ssgRNA with MS2 hairpin structure is:
[0060] ccggtgagaccgagagagggtctcagttttagagctaggccaacatgaggatcacccatgtctgcagggcctagcaagttaaaataaggctagtccgttatcaacttggccaacatgaggatcacccatgtctgcagggccaagtggcaccgagtcggtgctttttttt
[0061] All the above ssgRNA sequences were synthesized in Nanjing GenScript Biotechn...
Embodiment 2
[0081] Experimental design of embodiment 2 human TP53 gene point mutation repair
[0082] 1) The U-251 human glioma cell line was selected, and the sequencing results showed that the TP53 gene of this cell line carried a homozygous point mutation of R273H (CGT->CAT). To repair the point mutation, sgRNA (SaCas9) specifically targeting the TP53 gene was designed near this site ( Figure 6 ).
[0083] Its specific sequence:
[0084] agagaccggcgcacagaggaa
[0085] PAM: gagaat
[0086] 2) Designing a repair template: a DNA single oligonucleotide chain is selected as the template, and the sequence is actgggacggaacagctttgaggtgcgtgtttgtgcctgtcctgggcgtgataggcgaacggaagaggaaaacctccgcaagaaaggggagcctcaccacgagctgcccccaggggag. A synonymous mutation was introduced at the sgRNA recognition site of TP53 to avoid insertion / deletion mutations at the repaired site being cut by SaCas9.
[0087] 3) Transfect U251 cell line to verify mutation repair efficiency: In the experimental group, the SaC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com