A method for obtaining rdna ITS2 sequence for identification of Tiepi Fengdou
A tin foil mapper and sequence comparison technology, applied in DNA preparation, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems affecting DNA extraction efficiency, sample DNA degradation, low amplification efficiency, etc., and achieve easy mastery. , high accuracy, simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Identification of Example 1 Tiepi Fengdou
[0023] 1. Experimental materials
[0024] In this case, a total of 69 samples of Tiepi Fengdou were collected, and the specific information is shown in Table 1.
[0025] Table 1 Tiepi Fengdou sample information
[0026]
[0027]
[0028]
[0029] 2. Experimental equipment
[0030] Desktop high-speed centrifuge, PTC-100 gene amplification instrument, electrophoresis instrument, ultraviolet gel imaging analysis system, MM400 ball mill (Retsch, Germany).
[0031] 3. DNA extraction
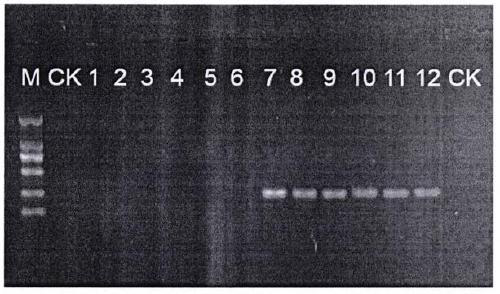
[0032] Take 20-30mg of Tiepi Fengdou dried medicinal materials, chop them up and crush them with MM400 ball mill, add 500-1000 μL of nuclear separation solution, mix well, centrifuge at 7000r / min for 5min, discard the supernatant, add 500-1000 μL of nuclear separation solution, Repeat this step 3-5 times until the supernatant is not viscous. The remaining steps were carried out according to the standard operating method of the Plant Geno...
Embodiment 2
[0049] The identification of embodiment 2 Dendrobium officinale base plant
[0050] Basically the same as Example 1, except that the original plant of Dendrobium candidum was used as the sample, DNA was extracted according to the above method, and the ITS2 sequence was amplified. As a result, the target sequence was also successfully obtained and the identification was completed.
Embodiment 3
[0051] The identification of embodiment 3 dendrobium officinale seed seedlings
[0052]Basically the same as Example 1, the difference is that Dendrobium officinale seeds and seedlings are used as samples, DNA is extracted according to the above method, and the ITS2 sequence is amplified. As a result, the target sequence can also be successfully obtained and the identification is completed.
[0053]
[0054]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com