MIRNA expression signature in classification of thyroid tumors
A technology of thyroid and expression profiling, which is applied in the direction of antineoplastic drugs, microbial determination/testing, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0366] Embodiment 1: the detection of microRNA in the sample of pre-operation
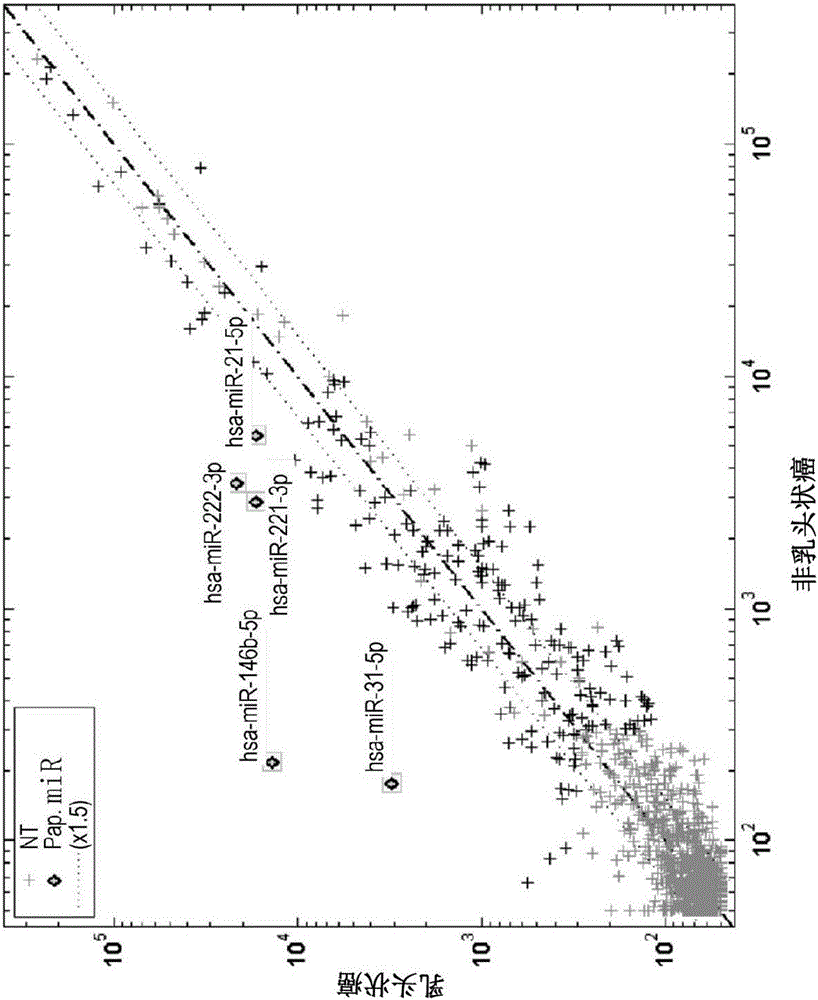
[0367] In several Papanicolaou, Giemsa and Diff-Quick stained smears from in vitro FNA biopsy samples, a preliminary study of microRNA profiling was performed to ensure the feasibility of the method. Since FNA smears usually have very few cells, providing very small amounts of RNA for research, such as 1 ng to 1000 ng, it is first necessary to evaluate whether microRNAs are detectable at such low RNA amounts. Thus, the microRNA expression levels of approximately 2200 individual microRNAs were measured in Giemsa-stained papillary and non-papillary carcinoma smears. In papillary carcinoma smears, five microRNAs (hsa-miR-146b-5p, hsa-miR-31-5p, hsa-miR-222-3p, hsa-miR- 221-3p and hsa-miR-21-5p) were overexpressed. figure 1 shows a comparison of microRNA expression between Giemsa-stained papillary carcinoma samples and non-papillary carcinoma samples, revealing microRNA signatures that are highly upr...
Embodiment 2
[0368] Example 2: Differential microRNA expression between malignant thyroid lesions and benign thyroid lesions
[0369] The sample cohort used in this experimental analysis consisted of 73 pre-manipulated thyroid FNA cell blockers selected from the archives of the Department of Pathology Temple University Hospital (Philadelphia, USA). The 73 samples included 35 samples of benign thyroid lesions and 38 samples of malignant thyroid lesions. The 35 benign tumors consisted of 18 samples of follicular adenoma, 8 samples of Hashimoto's thyroiditis, and 9 samples of hyperplasia (goiter). The 38 malignancies consisted of 10 follicular carcinomas and 28 papillary carcinomas. Of the 28 papillary carcinoma samples, 9 were papillary carcinoma, 13 were intracapsular follicular variant of papillary carcinoma, and 6 were non-intracapsular follicular variant of papillary carcinoma. Histological diagnosis ultimately assesses the malignancy or benignity of the thyroid lesion. Cytological cl...
Embodiment 3
[0378] Example 3: Distinguishing between different subtypes of malignant and benign thyroid lesions
[0379] Expression levels of miRs were compared between 18 follicular adenoma samples and 10 follicular carcinoma samples. Up- or down-regulated microRNAs in follicular adenoma versus follicular carcinoma are shown in Table 4.
[0380] Table 4: Up- or down-regulated miRNAs in follicular adenoma vs. up- or down-regulated miRNAs in follicular carcinoma
[0381]
[0382] P-values were calculated using a two-sided (unpaired) Student's t-test.
[0383] Fold change represents the proportion of the median value in each group.
[0384] AUC: Area under the curve when using miRNAs to classify two groups.
[0385] Median: The median (approximately) of the expressed values.
[0386] To compare miR expression levels in 18 follicular adenoma samples and 9 papillary carcinoma (non-follicular variant) samples, expression levels of hsa-miR-146b-5p and hsa-miR-21-5p were used to generat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com