PCR-Sort-PCR library enriching method applicable to high-throughput sequencing
A high-throughput, pcr-sort-pcr technology, applied in the field of high-throughput gene sequencing, can solve the problems of difficult access, difficulty in meeting the requirements of high-throughput sequencing library construction, and inability to develop low-input samples. Achieve the effects of easy practical implementation, solving the difficulty of library construction, and simple and feasible methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
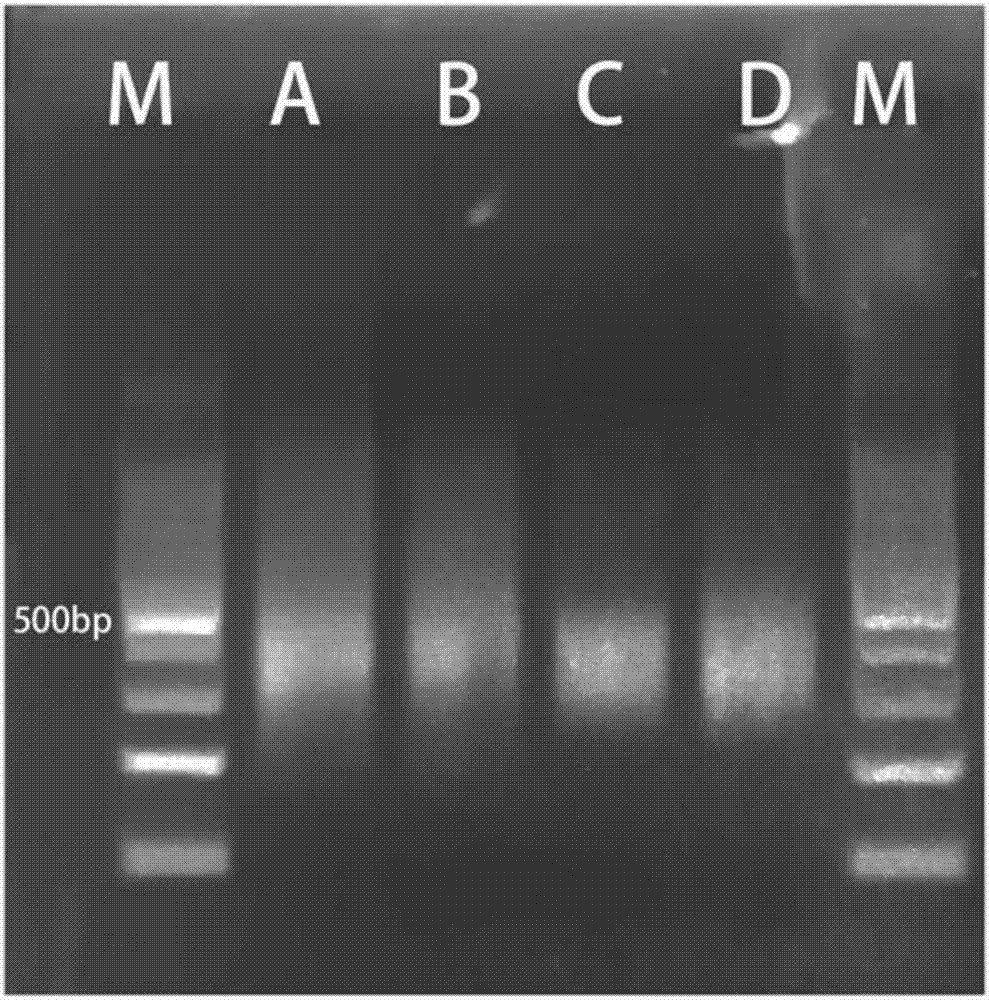
[0038] Example 1 Example of PCR-Sort-PCR (PSP) library enrichment technology applicable to high-throughput library construction of ultra-low starting nucleic acid templates of the present invention (1)
[0039] 1. Take the DNA extracted from the FFPE sample, the total amount is 5ng (not limited), and ultrasonically crush it to about 200bp;
[0040] 2. In the four PCR tubes A, B, C, and D, add 1 ng of DNA extracted from the crushed FFPE sample;
[0041] 3. According to the library construction kit VAHTS TM Universal DNA Library Prep Kit for (#ND604) provides the method and steps from library construction to ligation product purification (sample obtained after nucleic acid fragmentation, end repair, 3'end addition of A, adapter ligation, and ligation product purification);
[0042] 4. According to the PCR conditions given by the library construction kit, adjust the cycles to 9, and perform the first round of PCR reactions on samples C and D; directly perform PCR reactions on...
Embodiment 2
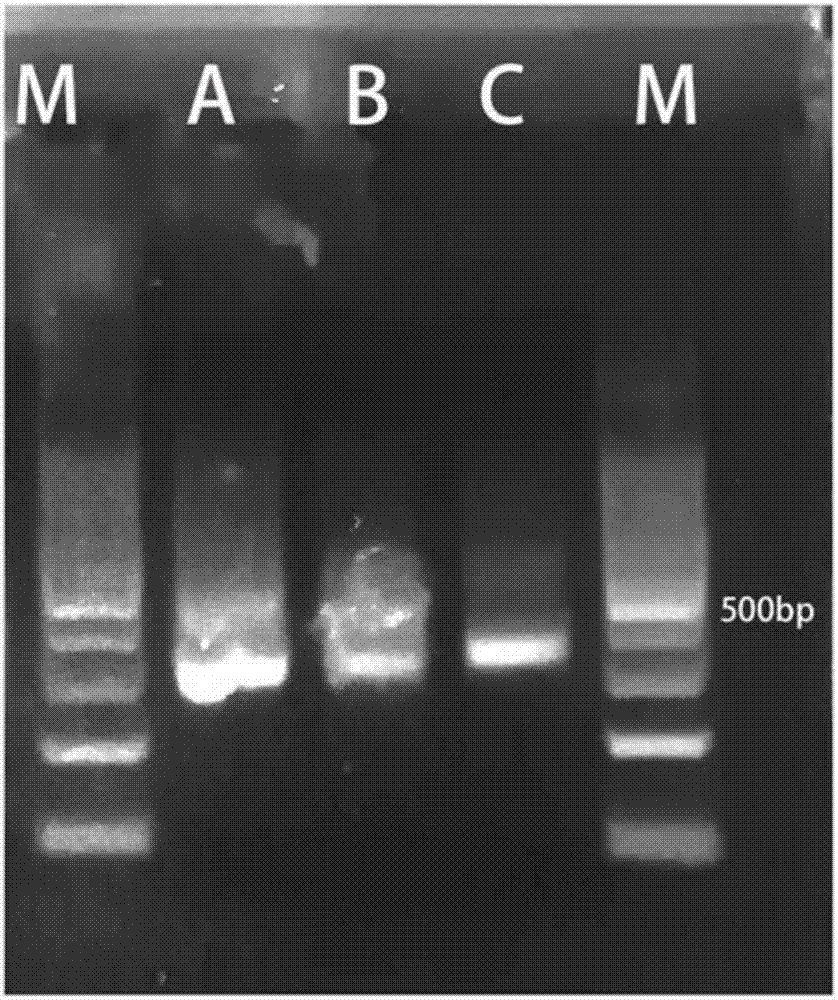
[0047] Example 2 Example of PCR-Sort-PCR (PSP) library enrichment technology applicable to high-throughput library construction of ultra-low starting nucleic acid templates of the present invention (2)
[0048] 1. Take the cfDNA sample, thaw it and put it on ice for later use;
[0049] 2. Add 1ng of cfDNA sample to each of the three PCR tubes A, B, and C;
[0050] 3. According to VAHTS TM Universal DNA Library Prep Kit for (#ND604) provides experimental methods and steps from library construction to ligation product purification;
[0051] 4. According to VAHTS TM Universal DNA Library Prep Kit for According to the PCR conditions given in (#ND604), adjust the cycles to 9, and perform the first round of PCR reaction for samples C; directly perform PCR reactions for 18 cycles for samples A and B;
[0052] 5. Sample A is purified by magnetic beads after the PCR reaction, sample B is subjected to fragment sorting; sample C is purified;
[0053] 6. Purification of the C sa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com