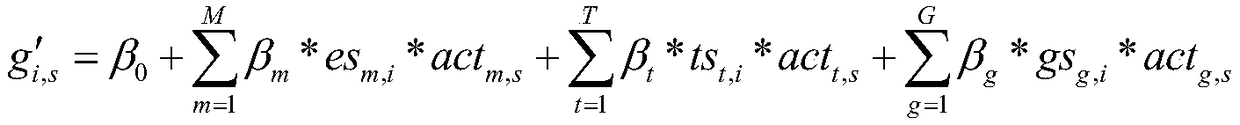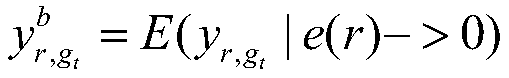A linear model-based method for identifying key regulators in gene co-regulatory networks
A technology of regulating network and linear model, applied in the field of computational biology, can solve the problems of high time complexity, failure to consider interaction and cooperative regulation relationship, and the recognition accuracy needs to be improved, so as to achieve the effect of improving accuracy and achieving simplicity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
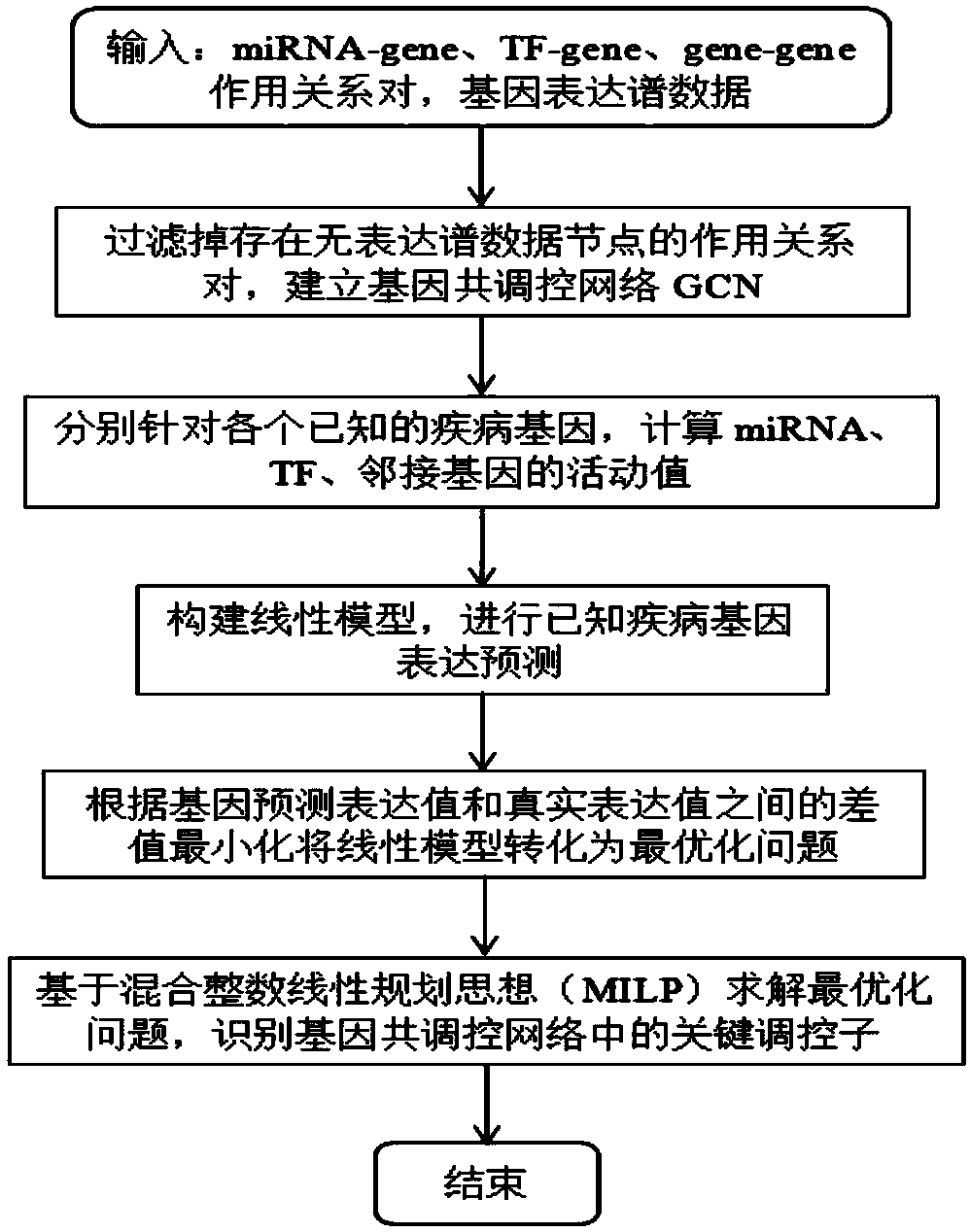
[0049] 1. Identification model of key regulators in gene co-regulatory network based on linear model
[0050] In the present invention, the key regulators in the gene co-regulation network are defined as: using gene expression profile data and gene regulation relationship, by constructing a linear model to predict the expression of known disease genes, thereby identifying the genes that seriously affect the disease in the co-regulation network Regulators of expression.
[0051] In order to clearly describe the key regulator identification model in the gene co-regulatory network based on the linear model, the inventors define the model as follows:
[0052] The proposed construct linear model predicts the expression of known disease genes in the form of:
[0053]
[0054] The goal of the linear model-based key regulator identification model in the gene co-regulatory network is to identify the regulators that seriously affect the expression of disease genes in the co-regulato...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com