A method for identification of hazelnut germplasm using nuclear its sequence
A germplasm and sequence technology, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve problems such as strong randomness, poor stability, limited sites, and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
[0023] The method of using the nuclear ITS sequence of the present invention to carry out the identification of hazel germplasm, its preferred embodiment is:
[0024] Amplification primers including a pair of Hazel ITS sequences:
[0025] Forward primer: 5'-AGTCGTAACAAGGTTTCCGT-3';
[0026] Reverse primer: 5'-TCCTCCGCTTATTTATATGC-3';
[0027] The method for carrying out the germplasm identification of hazelnut through the amplification primers of the above-mentioned pair of hazel ITS sequences, comprising the following steps:
[0028] Use forward primer and reverse primer to carry out PCR amplification on standard germplasm;
[0029] Use forward primer and reverse primer to carry out PCR amplification of the target germplasm to be identified;
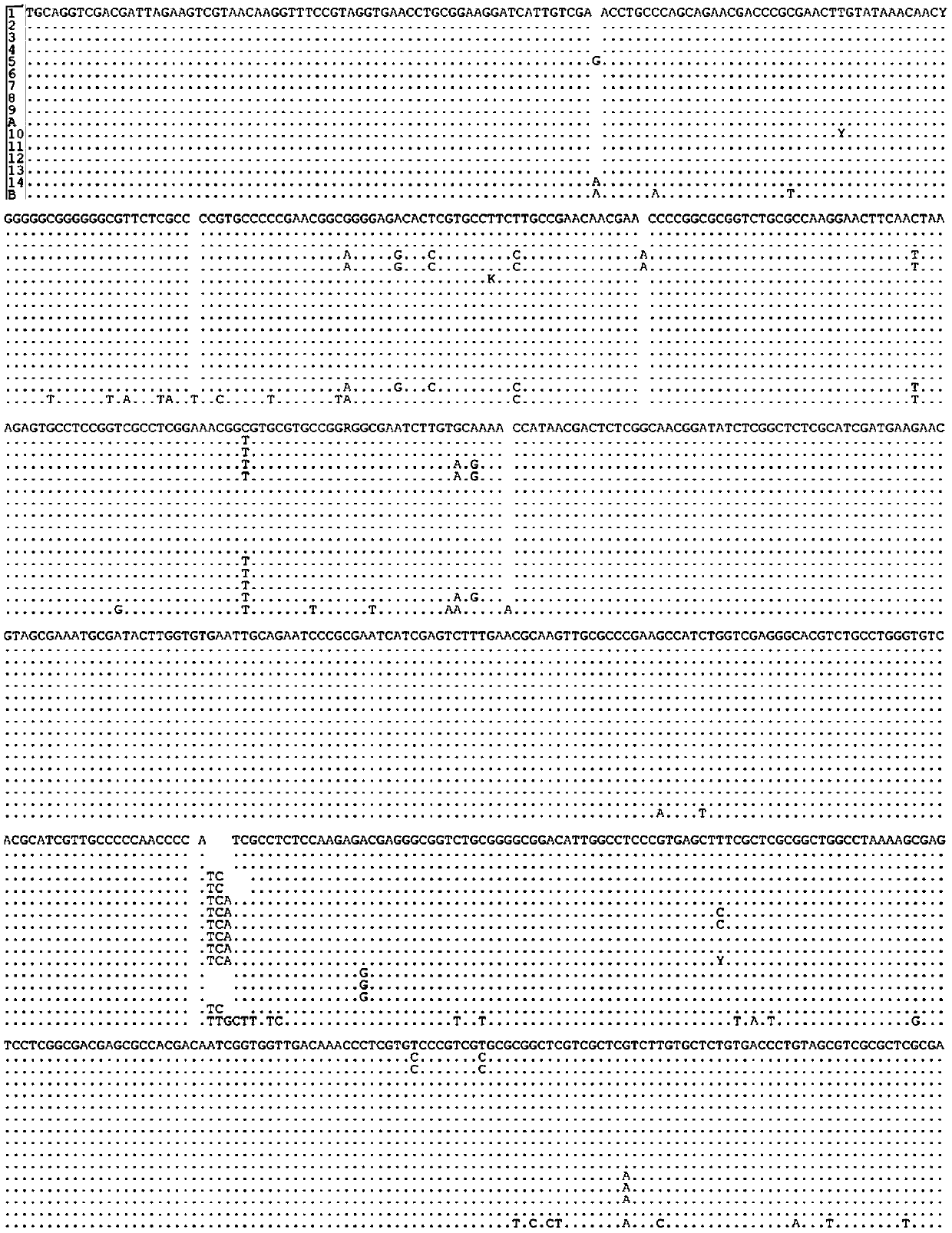
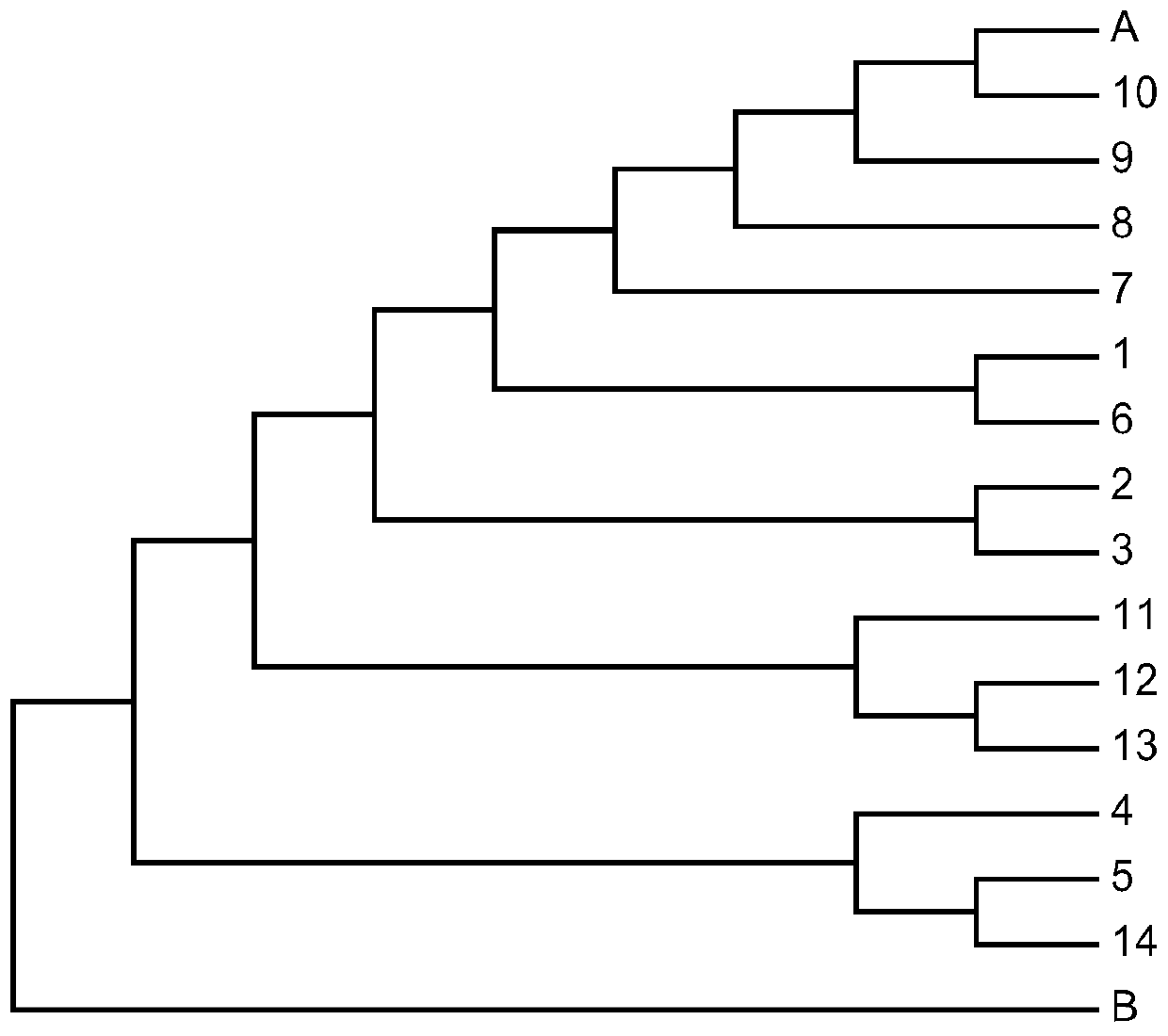
[0030] The amplified sequences obtained in the above two steps were compared, and the hazel germplasm identification was carried out according to whether the comparison results were consistent.
[0031] Before using the above-mentio...
no. 1 example
[0054] The first embodiment, the acquisition of Corylus plant material.
[0055] According to the actual survey, 14 Corylus plant materials were collected from different regions in my country, and the information of the 14 standard germplasm materials is shown in Table 1. A part of Corylus plant material A to be identified was collected from Taibai County, Shaanxi Province. The Qinling Mountains in Shaanxi Province are located in the cross-distribution area of Chuanzhen and Pingzhen. According to literature records, both Pingzhen and Chuanzhen are distributed in this area. According to traditional morphology, it is impossible to accurately identify which kind of Corylus the material to be identified is. In addition, there is another material collected from Chifeng, Inner Mongolia, whose shape is similar to that of hazel and tiger hazel, which is material B to be identified.
[0056] Table 1 Specific information of 14 Corylus plant materials
[0057]
no. 2 example
[0058] The second embodiment, the extraction of Corylus plant material genomic DNA
[0059] The optimized CTAB method was used to extract the whole genome DNA from the above 16 material samples, the specific method is as follows:
[0060] Step 201, preheating 2% CTAB extract and 2% β-mercaptoethanol in a 65°C water bath;
[0061] Step 202, take 0.2 g of leaves, add liquid nitrogen to fully grind, then quickly transfer to a 2 mL centrifuge tube, add 1 ml of preheated CTAB extract, bathe in 65 °C water for 60 min, and mix gently by inversion every 10 min;
[0062] Step 203, after the water bath is over, take out the centrifuge tube and cool to room temperature; centrifuge at 13000 rpm for 5 minutes, take 800 μL of the supernatant and transfer it to a new centrifuge tube;
[0063] Step 204, add 800 μL of chloroform / isoamyl alcohol (V:V=24:1), mix thoroughly, centrifuge at 13000 rpm for 5 min, and transfer the supernatant into a new 2 mL centrifuge tube;
[0064] Step 205, repea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com