A set of primers for detection of tussah microsporidia and their application
A technique for detecting microsporidia and primers, which is applied in the direction of microorganisms, microorganism-based methods, and microorganism measurement/inspection. Address technical limitations and reduce impact effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Preparation of plasmid standards:
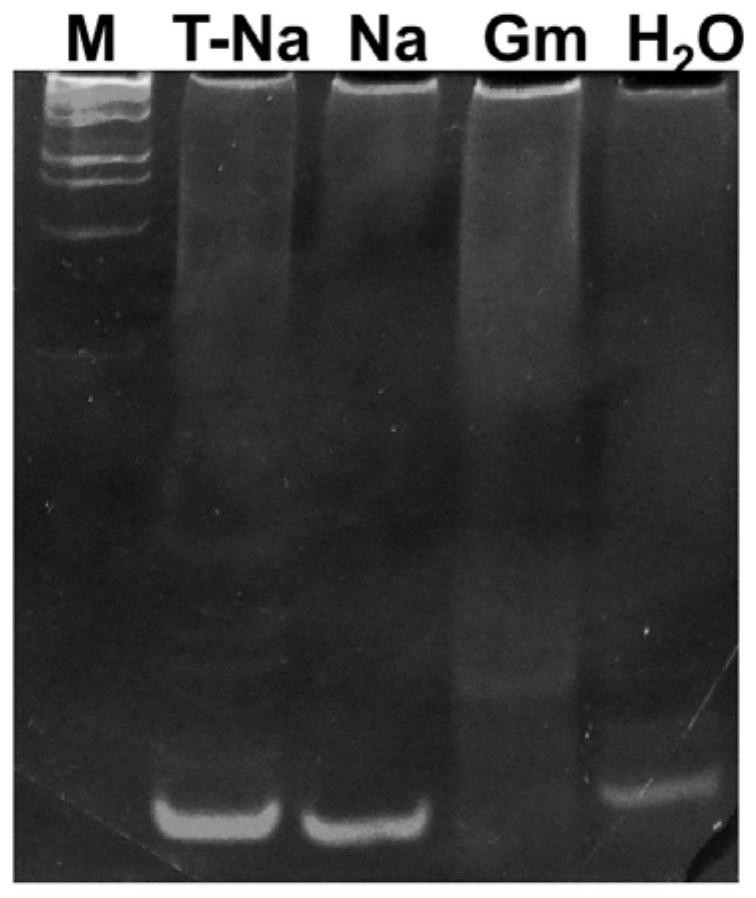
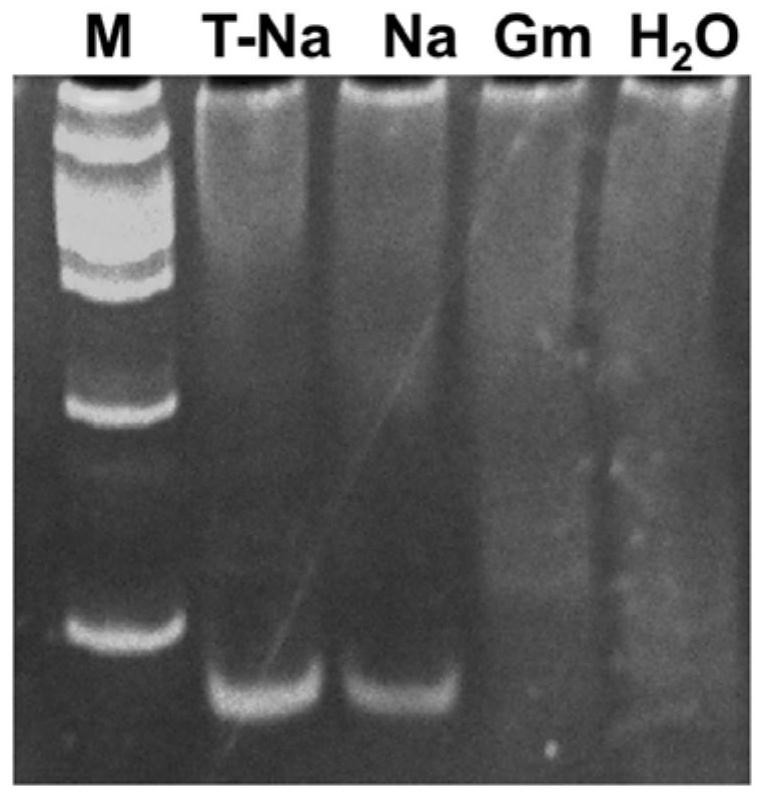
[0067] Take the tussah microsporidia suspension, centrifuge at 5000r / min for 5 minutes, remove the supernatant and retain the spore precipitate, add the extract (10mmol / L Tris-HCL pH 8.0; 10mmol / L EDTA pH 8.0; 100mmol / L NaCl; 2 %SDS; 0.039mol / L DTT), plus proteinase K (final concentration 100μg / mL), digested at 55°C for 4 hours, extracted with phenol / chloroform, washed twice with 70% ethanol, dried and eluted with TE to obtain Genomic DNA of Microsporidium tussah. The genomic DNA of the above-mentioned Microsporidia tussah was used for routine PCR. The PCR system was: 10×PCR buffer 2.5 μL, 10 mmol dNTPs 2 μL, MgCl2 2 μL, 10 μmol / L upstream and downstream primers 0.5 μL each, 1U Taq enzyme, template DNA 2 μL, add Sterile deionized water to 25 μL. Reaction conditions: pre-denaturation at 94°C for 5 minutes; 35 cycles at 94°C for 30s, 65°C for 30s, and 72°C for 60s; extension at 72°C for 10 minutes. After the reaction, take 10 μL of t...
Embodiment 2
[0079] Preparation of plasmid standards:
[0080]Take the tussah microsporidia suspension, centrifuge at 5000r / min for 5 minutes, remove the supernatant and retain the spore precipitate, add the extract (10mmol / L Tris-HCL pH 8.0; 10mmol / L EDTA pH 8.0; 100mmol / L NaCl; 2 %SDS; 0.039mol / L DTT), plus proteinase K (final concentration 100μg / mL), digested at 55°C for 4 hours, extracted with phenol / chloroform, washed twice with 70% ethanol, dried and eluted with TE to obtain Genomic DNA of Microsporidium tussah. The genomic DNA of the above-mentioned Microsporidia tussah was used for routine PCR. The PCR system was: 10×PCR buffer 2.5 μL, 10 mmol dNTPs 2 μL, MgCl2 2 μL, 10 μmol / L upstream and downstream primers 0.5 μL each, 1U Taq enzyme, template DNA 2 μL, add Sterile deionized water to 25 μL. Reaction conditions: pre-denaturation at 94°C for 5 minutes; 35 cycles at 94°C for 30s, 65°C for 30s, and 72°C for 60s; extension at 72°C for 10 minutes. After the reaction, take 10 μL of th...
Embodiment 3
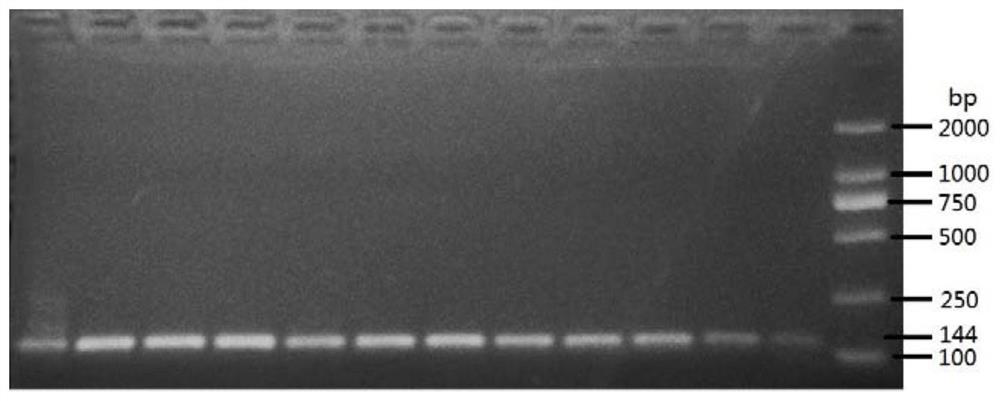
[0092] The DNA of Microsporidium tussah was automatically extracted by Thermo Nucleic Acid Automatic Extractor 706. A little tissue from 12 pupae with microparticle disease was taken, soaked in 210 μL of lysate (0.1M NaOH, 1% SDS) for 2 hours, and 200 μL of the supernatant was removed for use. Use the KF_TissueDNA_Duo program preset by the instrument to slightly adjust the amount of solvent in the DW wells of column A: take 200 μL of the lysate that has fully lysed the sample, 20 μL of the magnetic bead suspension (sigma), and 140 μL of absolute ethanol. Take 2 μL of the extracted DNA as a PCR template to configure a reaction system: each 25 μL system contains 0.5 μL of each primer (10 pM) shown in SEQ ID NO:4 and SEQ ID NO:5, 0.25 μL of rTaq, 2.5 μL of 10×Buffer, 2 μL of dNTPs, 17.25 μL of deionized water and 2 μL of DNA template. The PCR reaction conditions are 95°C for 4 minutes; 40 cycles of 95°C for 30 seconds, 65°C for 30 seconds, and 72°C for 1 minute, and then extende...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com