Prediction method and system of protein local structure characteristics
A local structure and prediction method technology, applied in the field of bioinformatics, can solve the problems of high cost and low efficiency of protein tertiary structure, and achieve the effect of solving high cost, low efficiency and improving overall performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
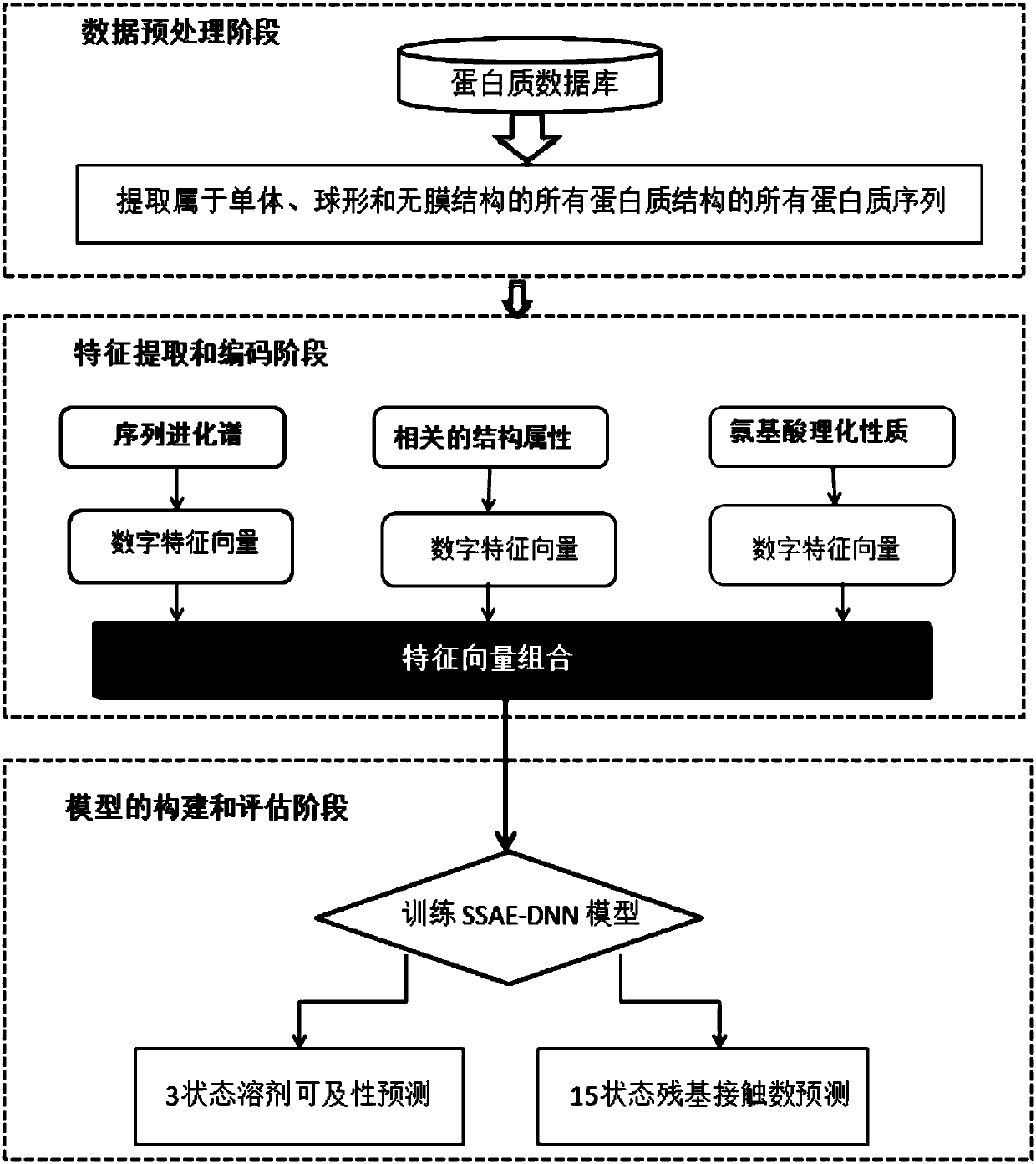
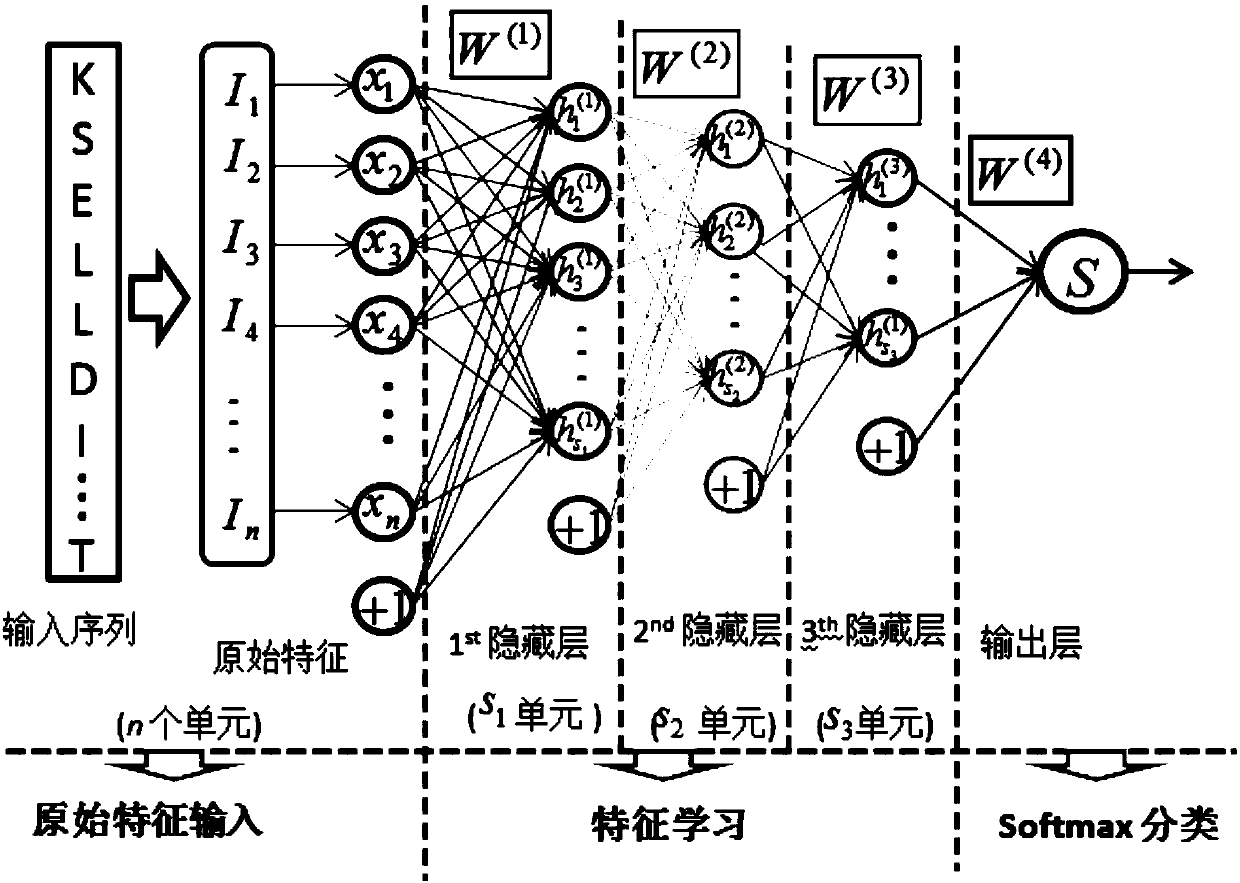
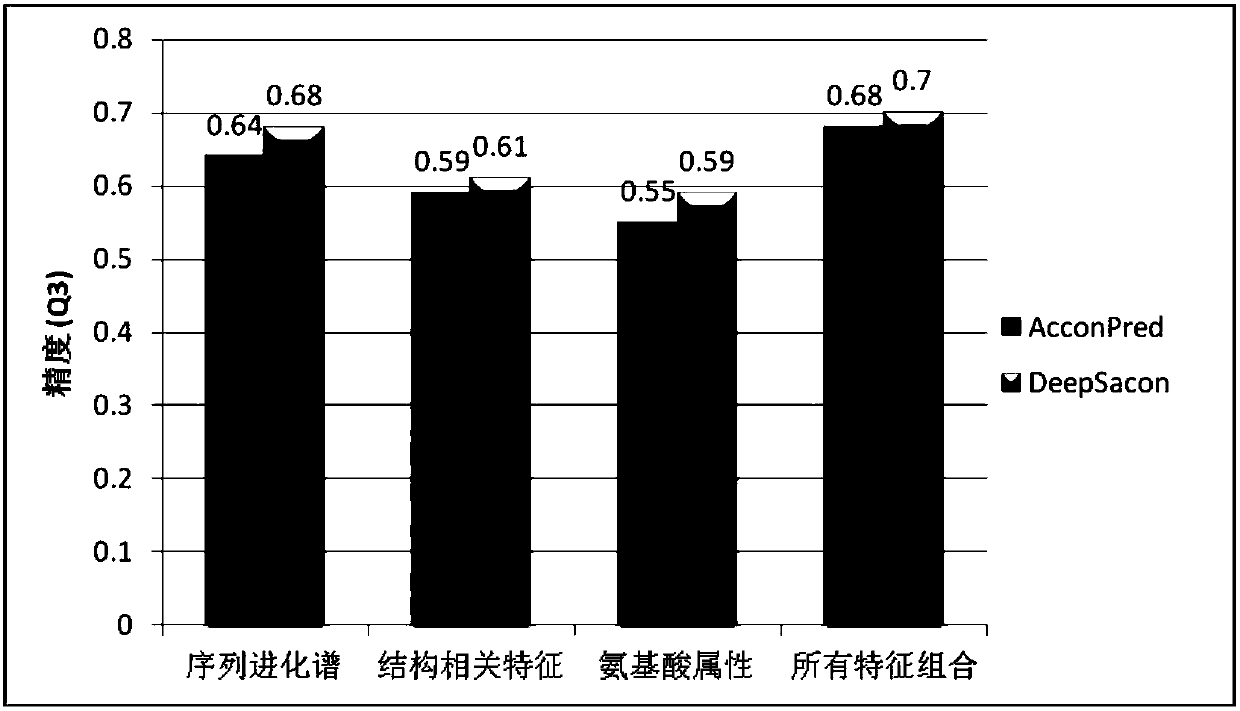
[0047] This embodiment discloses a method for predicting local structural features of proteins, referring to figure 1 , the first is the data preparation stage, extracting all protein sequences belonging to monomer, spherical, and non-membrane structures from the protein database to form a training data set. Next is the feature encoding stage, which is to convert the character strings in the protein sequence text into numerical features, and different features can be encoded by different software and programs. The present invention divides all original features into three categories: sequence evolution spectrum, The relevant structural properties and amino acid physicochemical properties are predicted, and then all the features are combined together as the original input of the model. Finally, it is the training and prediction stage of the model. The value encoded in the second stage is used as input to train the stacked sparse self-encoder neural network (SSAE-DNN). For a gi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com