Application of lncRNA-NEAT1 as molecular pathological diagnosis marker in preparation of glioma diagnosis reagent
A technology of pathological diagnosis and diagnostic reagents, which is applied in the field of medicine, can solve the problems of high price, poor sensitivity, and long experiment cycle, and achieve the effect of low cost, high sensitivity, and time period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Screening and identification of lncRNA-NEAT1
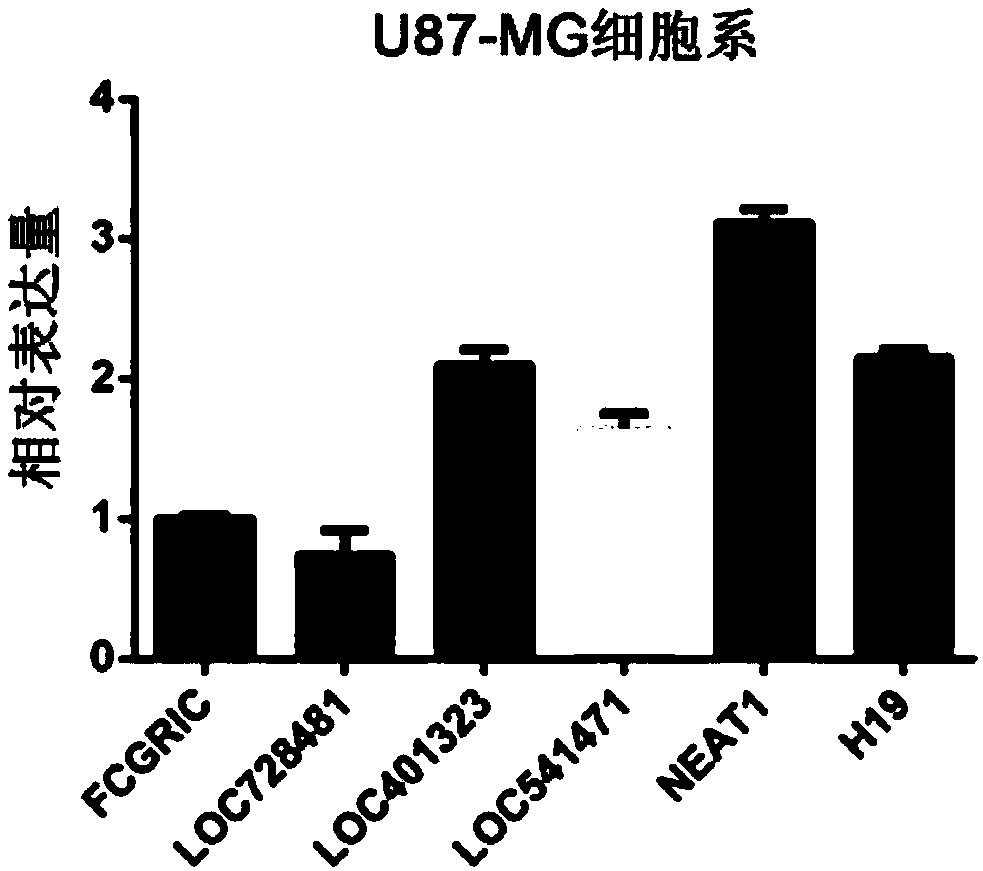
[0035] The inventors of the present invention screened 202 common lncRNAs from the accumulated 1500 cases of glioma databases CGGA, Rembrandt, TCGA and GSE16011, and found that 6 lncRNAs were highly expressed in glioma EGFR module type, and in the above-mentioned The 6 lncRNAs were verified by PCR in the U87-MG cell line in the 4 major glioma authoritative public databases, and it was found that the expression of lncRNA-NEAT1 was the highest in the U87-MG cell line (see figure 1 ).
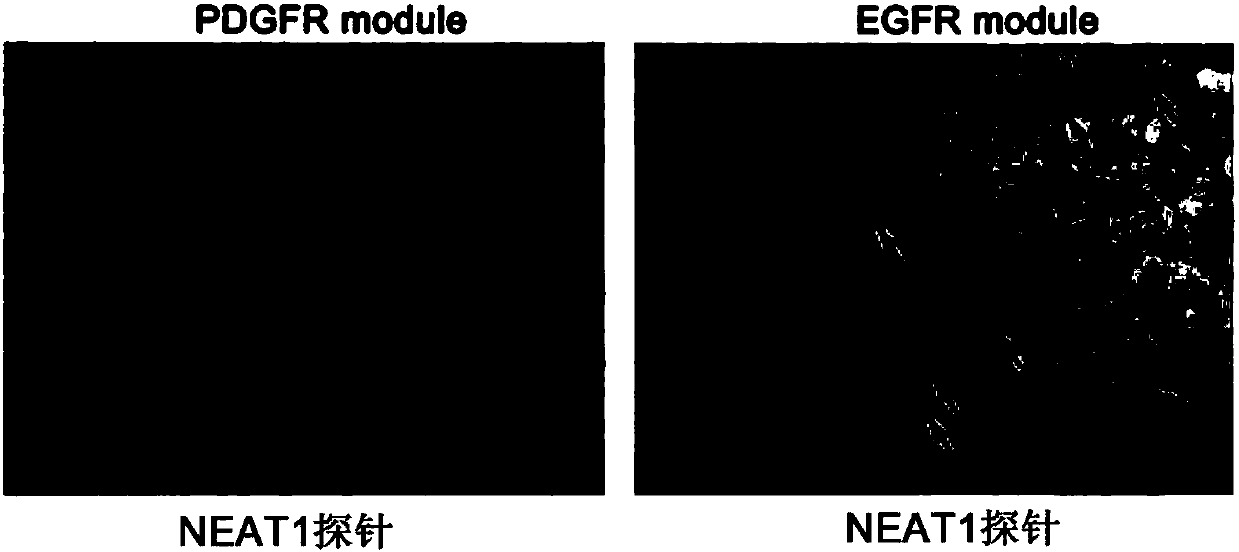
[0036] In situ hybridization was used to detect the expression level of lncRNA-NEAT1 in glioma tissues of patients with PDGFR module and EGFR module, and the results were as follows figure 2 shown.
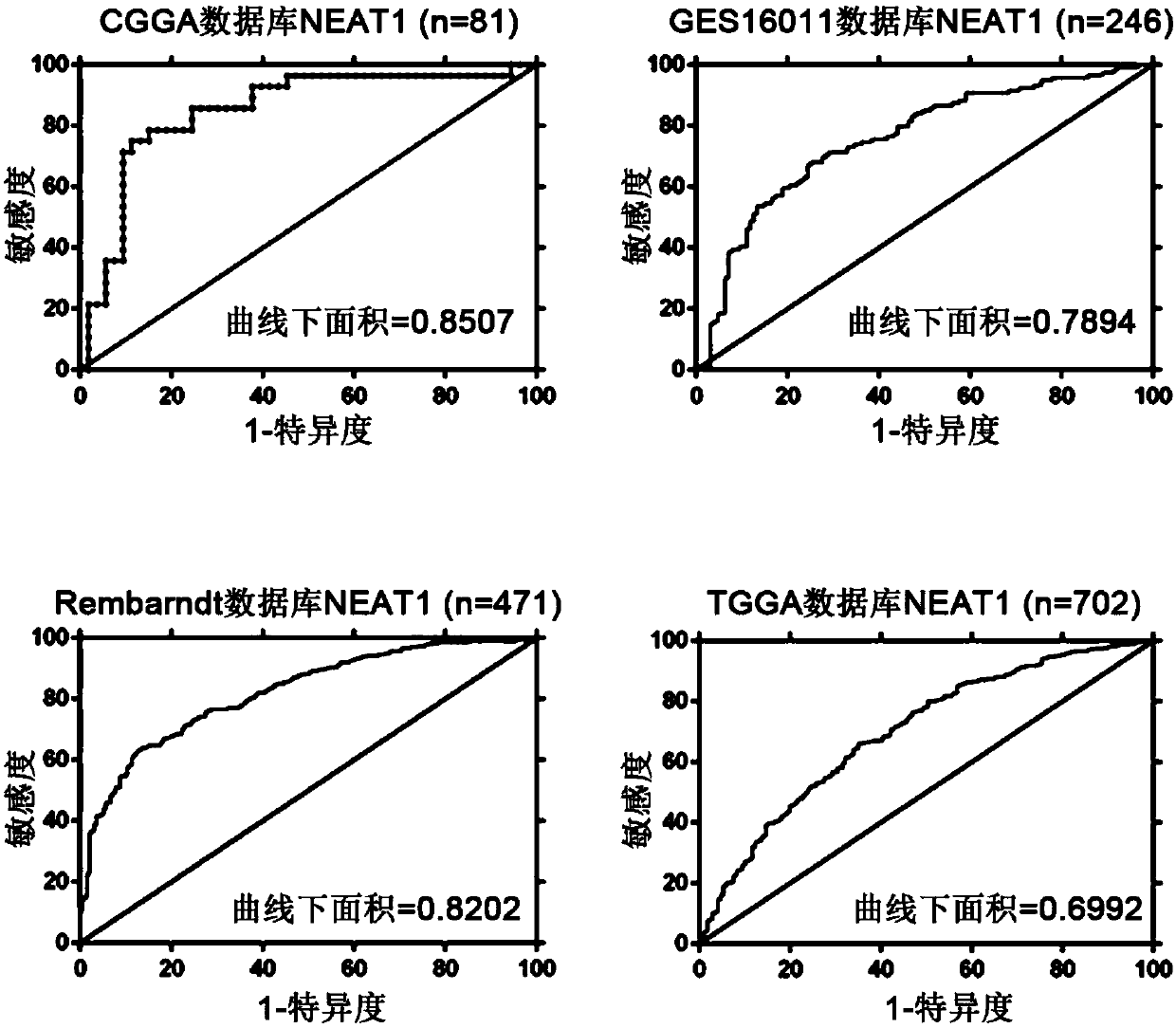
[0037] Then, the receiver operating characteristic curve (ROC) of NEAT1 in the EGFR module was evaluated in the above four authoritative public databases. The results are as follows: image 3 shown.
[0038] These results show that l...
Embodiment 2
[0039] Example 2 Application of lncRNA-NEAT1 as a molecular pathological diagnostic marker in the detection of EGFR module glioma
[0040] 1. Collection, processing and extraction of RNA from tumor tissue samples
[0041] (1) Collection of glioma tissue samples: pathological tissues of glioma patients were collected.
[0042] (2) Treatment of glioma tissue samples: Rinse with PBS and store in liquid nitrogen.
[0043] (3) Extraction of RNA in glioma tissue: 100 mg of each tissue sample was taken, ground into powder, and RNA was extracted according to Trizol purchased from Sigma Company.
[0044] (4) Reverse transcription into cDNA: take the same amount of RNA from each sample, and perform reverse transcription with random primers. According to 5×M-MLVRT Buffer 5μl, MgCl 2 2 μl, M-MLV Reverse transcript 1 μl, rRNasin 0.5 μl (Promega), dNTPs 1 μl, random primer 1 μl, extracted RNA 2ul, DEPC water (supplemented with H 2 O to bring the total volume to 20 μl) for reverse trans...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com