Brassica napus seed number per silique major gene locus close linkage molecular markers and applications thereof
A technology of molecular markers and rapeseed, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem of ineffective discovery and utilization of favorable genes of germplasm resources, lack of independent intellectual property rights and breeding value genes and labeling, poor selection effect of complex quantitative traits, etc., to achieve the effect of convenient and fast detection method, accurate and fast screening, and saving production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
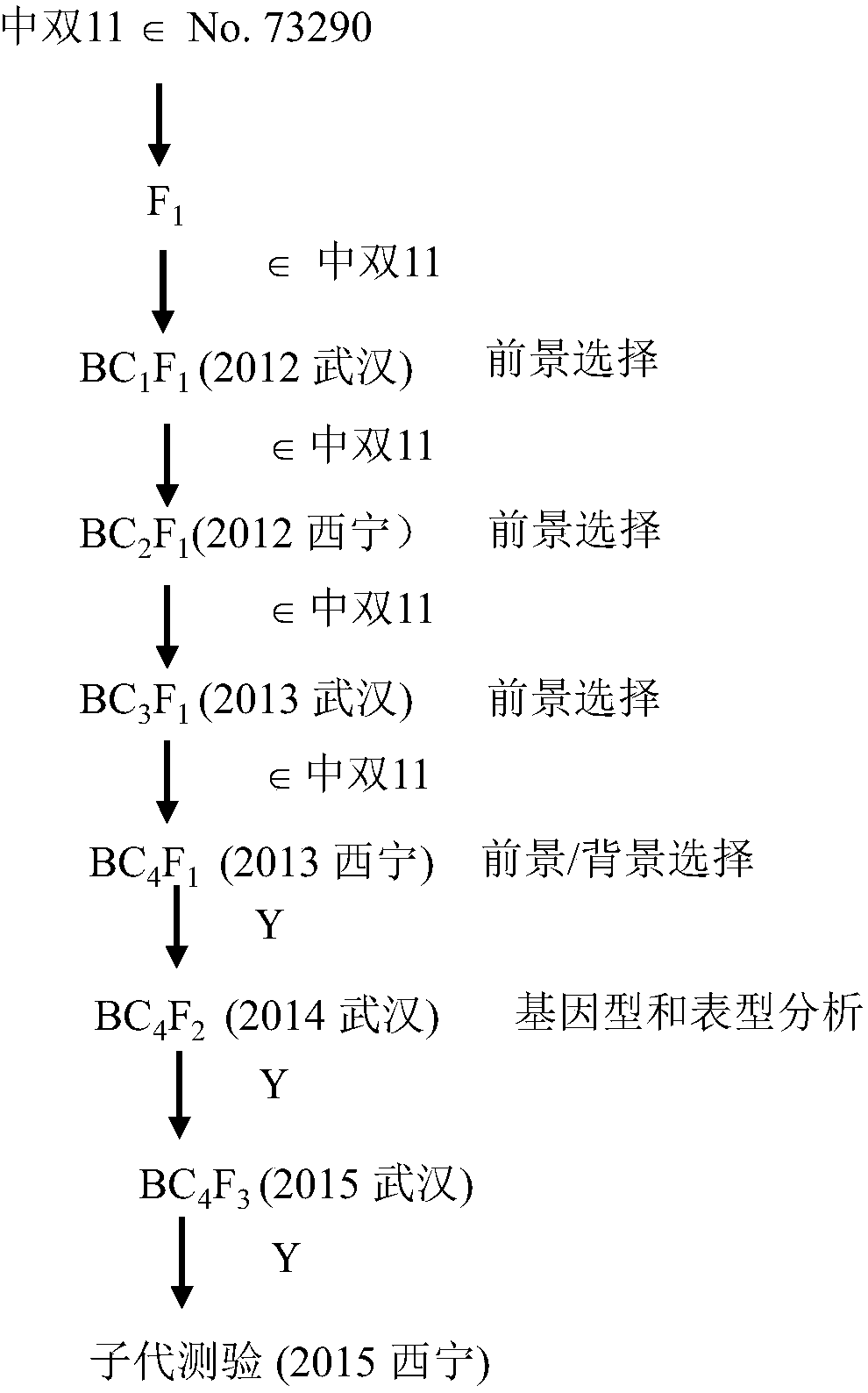
[0025] Embodiment 1: Construction of QTL-NIL
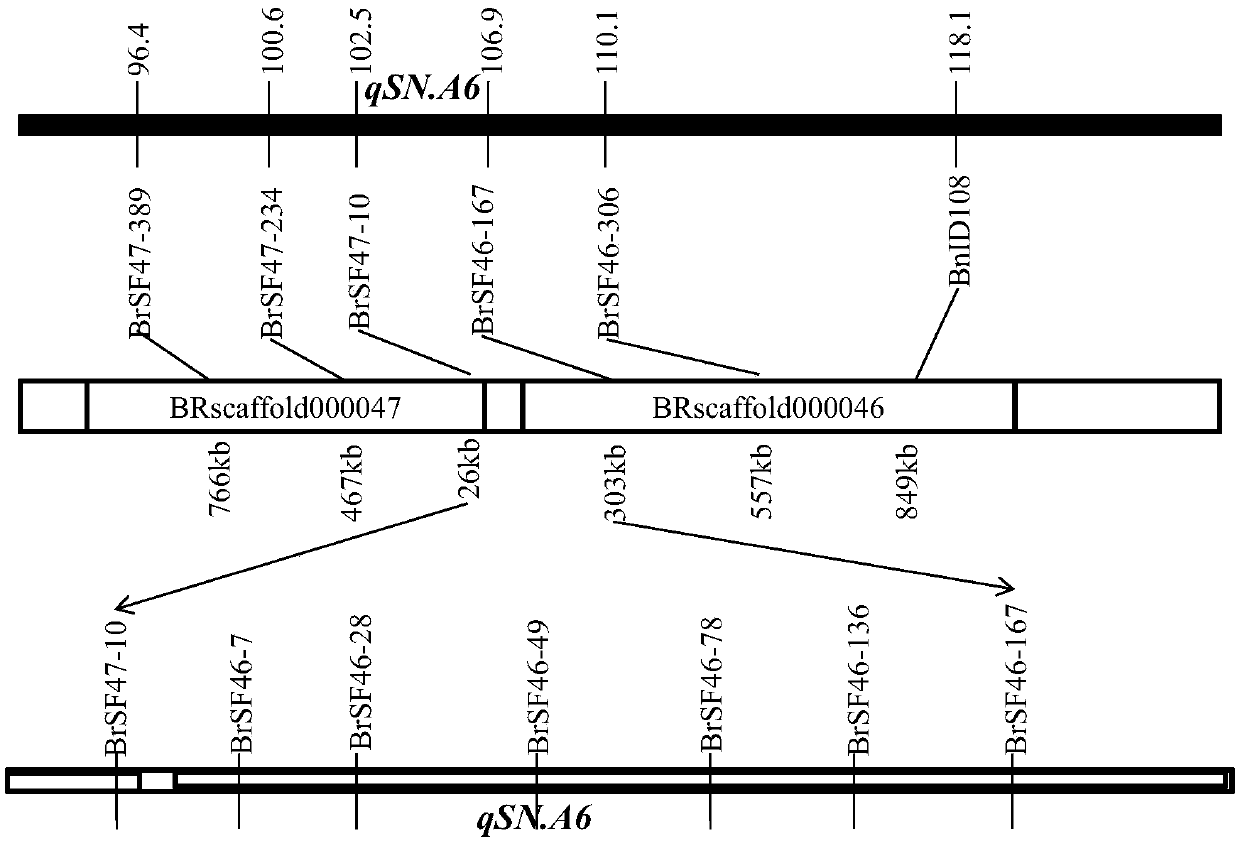
[0026] According to the strategy of fine positioning ( figure 1 ), NIL was constructed against qSN.A6. In this experiment, the hybrid F 1 During this period, the molecular markers BrSF47-389 and BnID108 on both sides of qSN.A6 were used to select heterozygous individual plants and double 11 consecutive backcrosses in the recurrent parents for 4 generations to obtain BC 4 f 1 generation. Then, 80 molecular markers (Yang et al., 2016) other than the target main effect QTL and evenly distributed on the 19 linkage groups of rapeseed were used for genetic background screening, from BC 4 f 1 In the population, select a single plant that is heterozygous for the target fragment and has a background recovery rate >95% to self-cross to obtain BC 4 f 2 seed. Field Planting BC 4 f 2 Generation of QTL-NIL segregation populations, using the above molecular markers to screen recombinant individual plants, and harvest test species at th...
Embodiment 2
[0027] Example 2: Development of Molecular Markers
[0028] First, use self-designed primers to compare with the genome sequence of Chinese cabbage or Brassica napus to determine the genomic region corresponding to the target QTL. This genomic region was searched for SSRs using MISA software. Using BWA software, the parental No.73290 resequencing sequence was mapped to the reference genome sequence of Shuang11 in the parent, and the InDel site of the target QTL interval was found by samtools software. Then, use Primer3.0 software to design SSR / InDel primers.
[0029] Using the magnetic bead method to extract the genomic DNA of the leaves of Shuang11 and No.73290 and QTL-NIL populations in the parents, using the self-developed SSR / InDel primers, and PCR amplification of the parental DNA, the products were in denaturing polyacrylamide gel After electrophoresis, staining and developing, the size of the bands was discriminated, and polymorphic primers were screened.
[0030] PC...
Embodiment 3
[0036] Example 3: Genotype analysis of QTL-NIL segregation population and screening of recombinant individual plants
[0037] (1) Using the magnetic bead method to extract the genomic DNA of the QTL-NIL isolated population;
[0038] (2) Using the molecular markers developed above to carry out molecular marker analysis on the genomic DNA of the QTL-NIL segregation population;
[0039] (3) Analyze the genotypes of these molecular markers in the QTL-NIL segregation population and screen recombinant individual plants.
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap