A method for constructing a large fragment library and its application
A construction method and large fragment technology, applied in the field of molecular biology, can solve the problems of inability to specifically digest DNA fragments, high manpower reagents and reagent storage costs, read length, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Embodiment 1 large fragment DNA library construction
[0068] 1. Large fragment processing of DNA samples
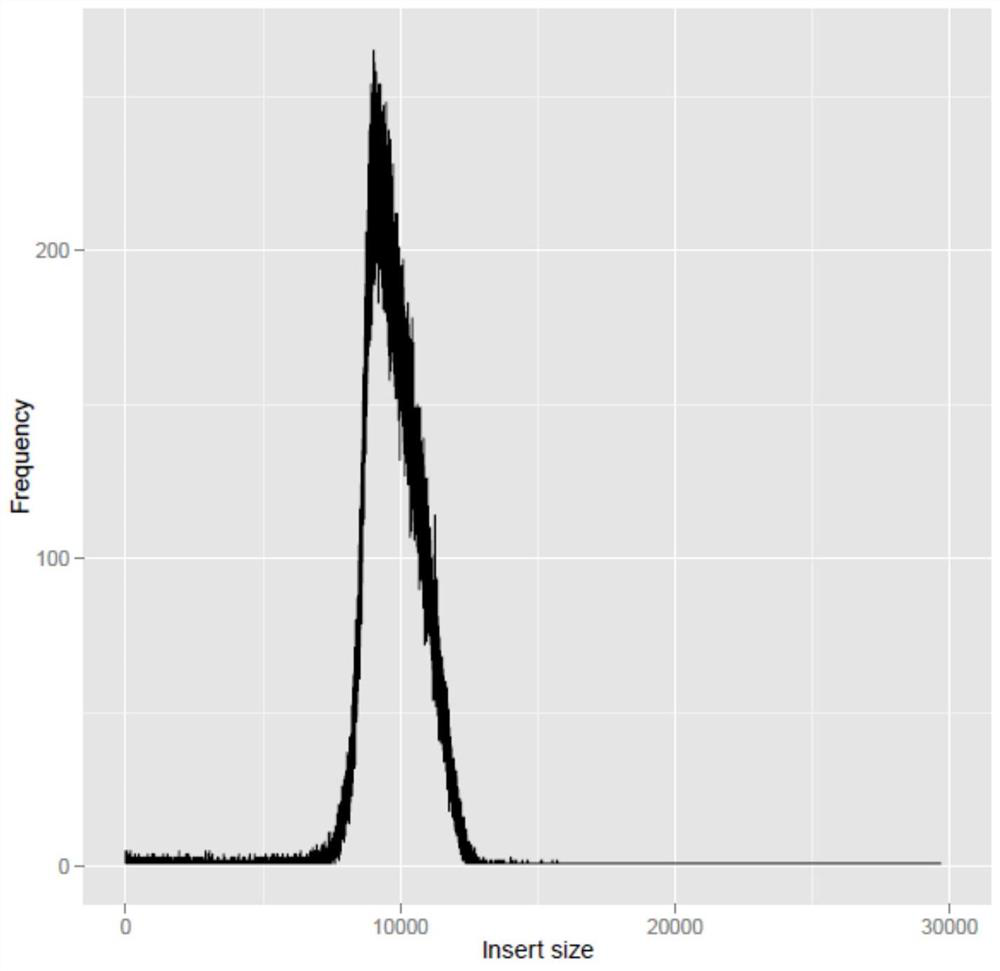
[0069] 1.1 Take 2mL whole blood from healthy people and extract according to the operating instructions of the blood genome DNA extraction system (0.1-20mL) (Tiangen Biochemical Technology Co., Ltd.) to obtain DNA samples. Take 10 μg of DNA sample, and use the HydroShearPlus DNA Fragmentation Instrument to fragment large fragments of DNA samples according to the instructions. The target large fragment length is 10kb.
[0070] 1.2 Prepare 0.8% agarose gel, use TIANGEN company 1Kb DNA Ladder as the molecular weight standard, and electrophoresis at 100V for 2 hours. After electrophoresis, the gel was taken out and stained in TAE containing EB dye for 20 minutes. Fragments around 9-12kb were cut and glued under ultraviolet irradiation.
[0071] 1.3 Put the excised gel into a weighed clean 2.0mL centrifuge tube, and use the agarose gel extraction kit to perform gel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com