Method for automatically analyzing GC-MS (Gas Chromatography-Mass Spectrometry) overlapping peak to accurately recognize compounds
An accurate identification and automatic analysis technology, applied in the field of analyzing gas chromatography-mass spectrometry data, can solve the problems of inability to promote non-targeted metabolomics, automatic data analysis, and difficulty in overlapping signal analysis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0026] In combination with the accompanying drawings of the present invention, the technical solutions of the embodiments of the invention will be further elaborated in detail.
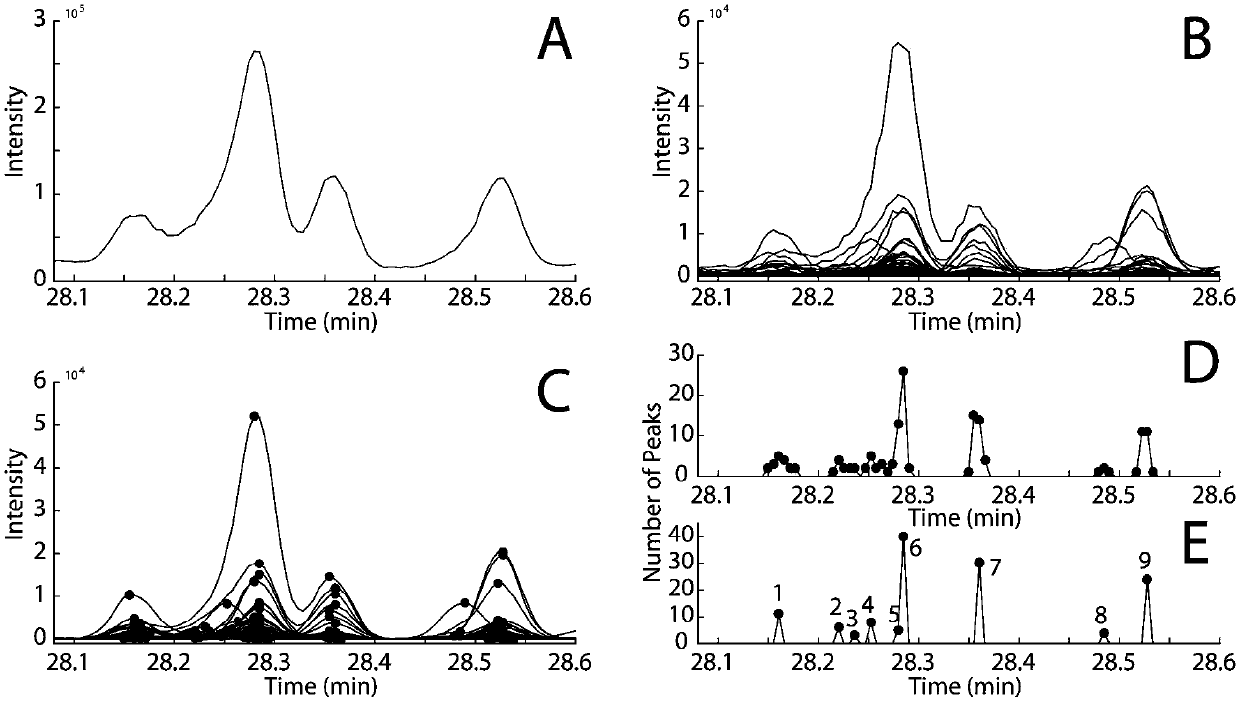
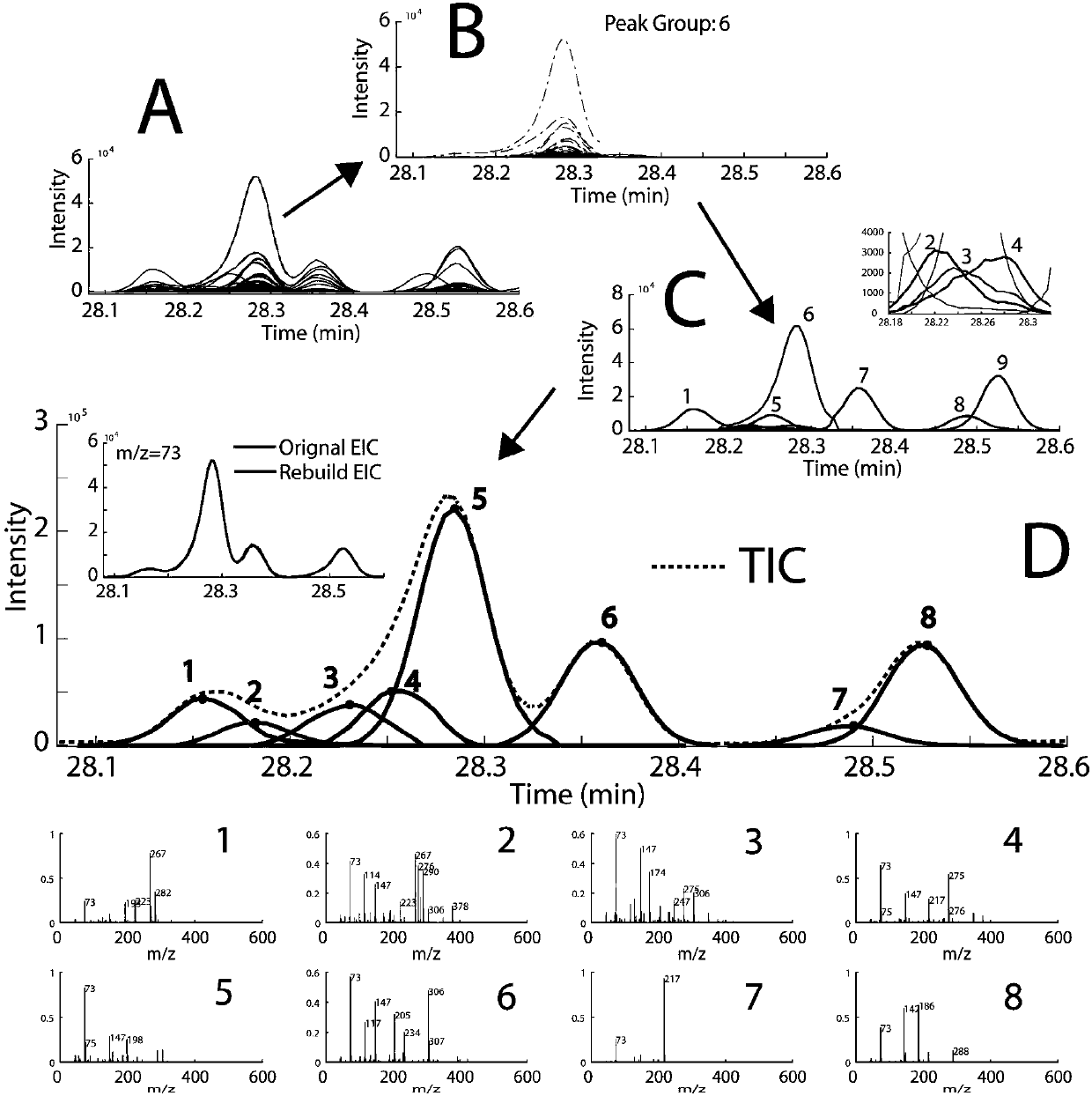
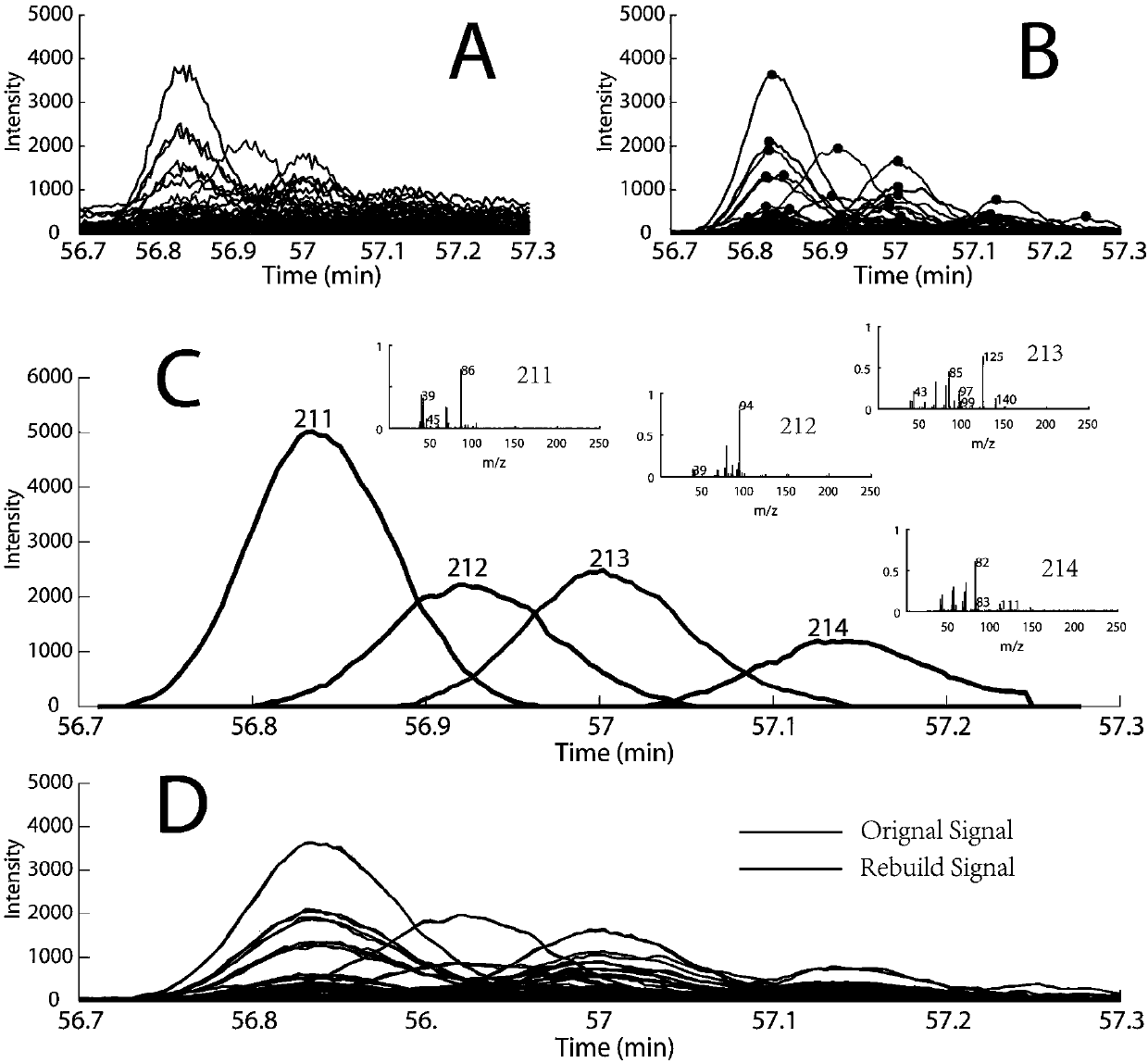
[0027] The present invention demonstrates the whole process of the method for automatically analyzing GC-MS overlapping peaks and accurately identifying compounds through the following examples.
[0028] S1: Plant sample pretreatment:
[0029] Tobacco leaf samples were freeze-dried and pulverized. Weigh 100 mg of the sample, extract with 5 mL of dichloromethane, vortex, sonicate and centrifuge, take 1 mL of the supernatant, dry it, add 100 μL of BSTFA, and derivatize at 70 ° C for 1 h. After GC-MS analysis, the compound data information after single-sample GC-MS analysis is obtained. The GC-MS analysis conditions are as follows: Chromatographic conditions: the chromatographic column is an Agilent DB-5MS column (60m×2.5mm, i.d., 2.5μm). The temperature was programmed to rise, the initial column temp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com