A eukaryotic expression vector expressing circular RNA
An overexpression vector, eukaryotic technology, applied in DNA/RNA fragmentation, recombinant DNA technology, introduction of foreign genetic material using vectors, etc., can solve the problem that circRNA has not been discovered and has not been studied yet, and achieves stable results and easy operation. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
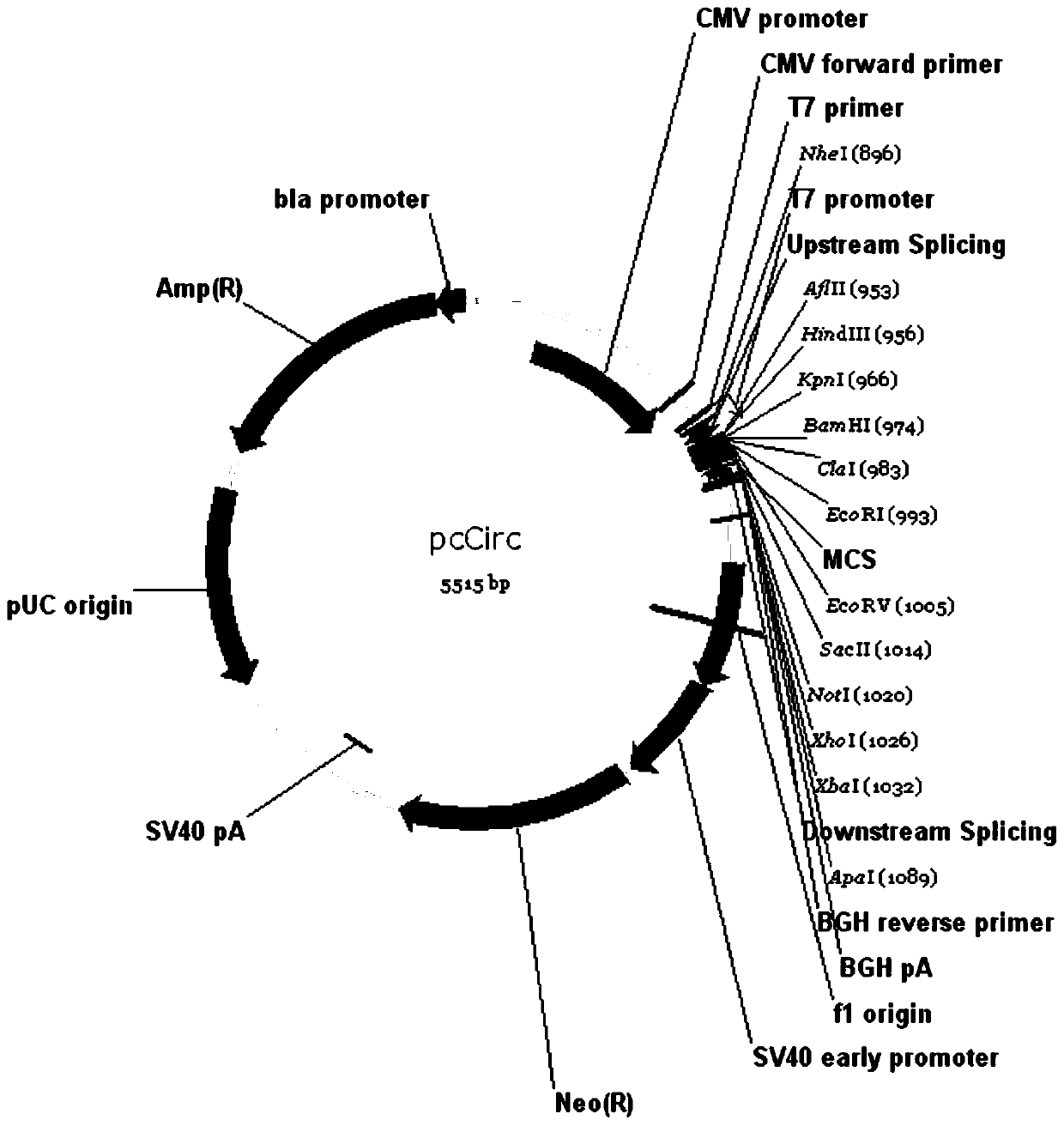
[0059] Embodiment 1, the construction of the plasmid of pcCirc
[0060] The technologies involved in the present invention are conventional technical means of molecular cloning, and the enzymes, primers, reagents and reaction conditions involved in them can be reasonably selected according to the experience of those skilled in the art unless otherwise specified, and the reagent consumables involved belong to Commercially available common products, detection means and instruments involved therein are also well known and mastered by those skilled in the art. The technical solution of the present invention will be further described through examples and test examples, but it should not be construed as a limitation of the present invention.
[0061] Taking the commercial expression vector (pcDNA3.1) as an example, we constructed the overexpression vector pcCirc, in which the sequence of the multiple cloning site region is:
[0062] CTTAAGCTTGGTACCGAGCTCGGATCCACATCGATTGGTGGAATTCTGC...
Embodiment 2
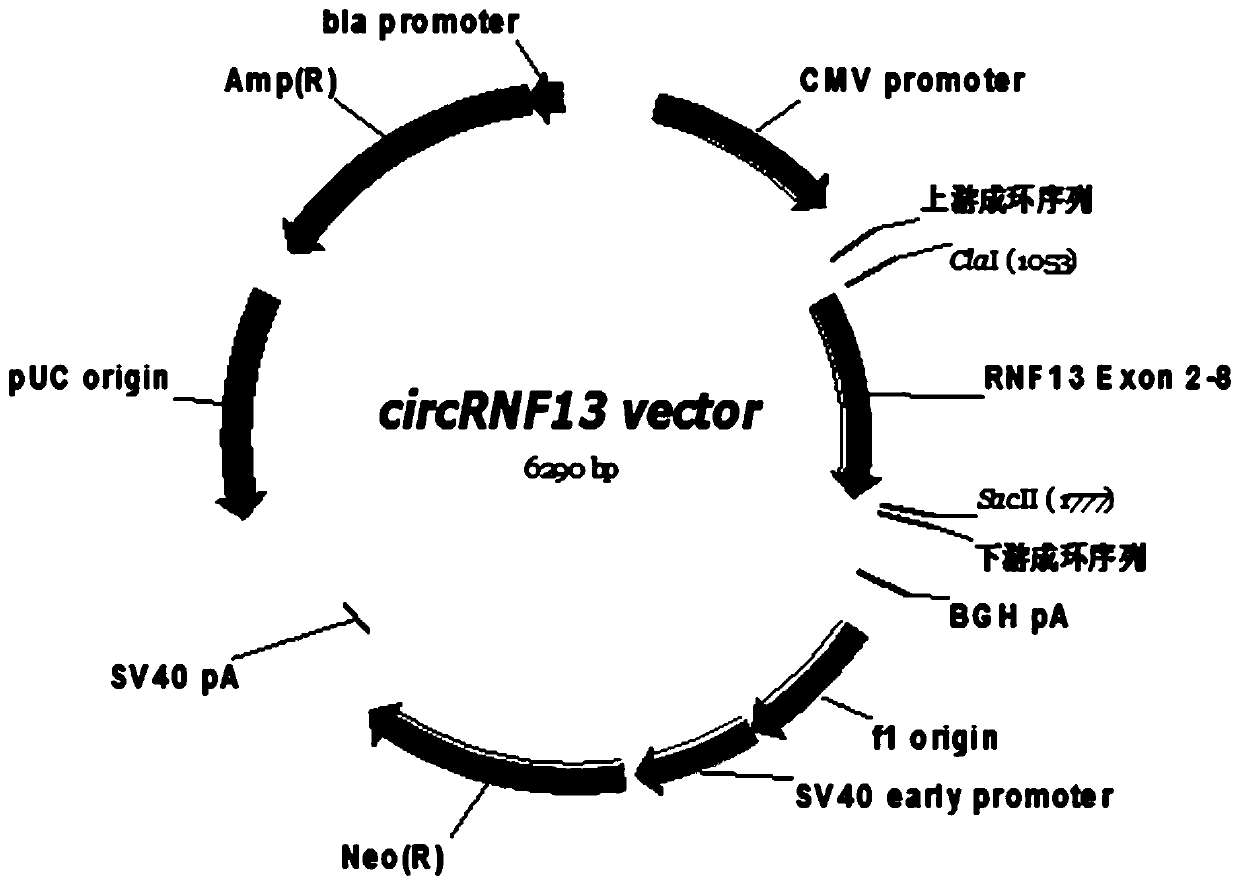
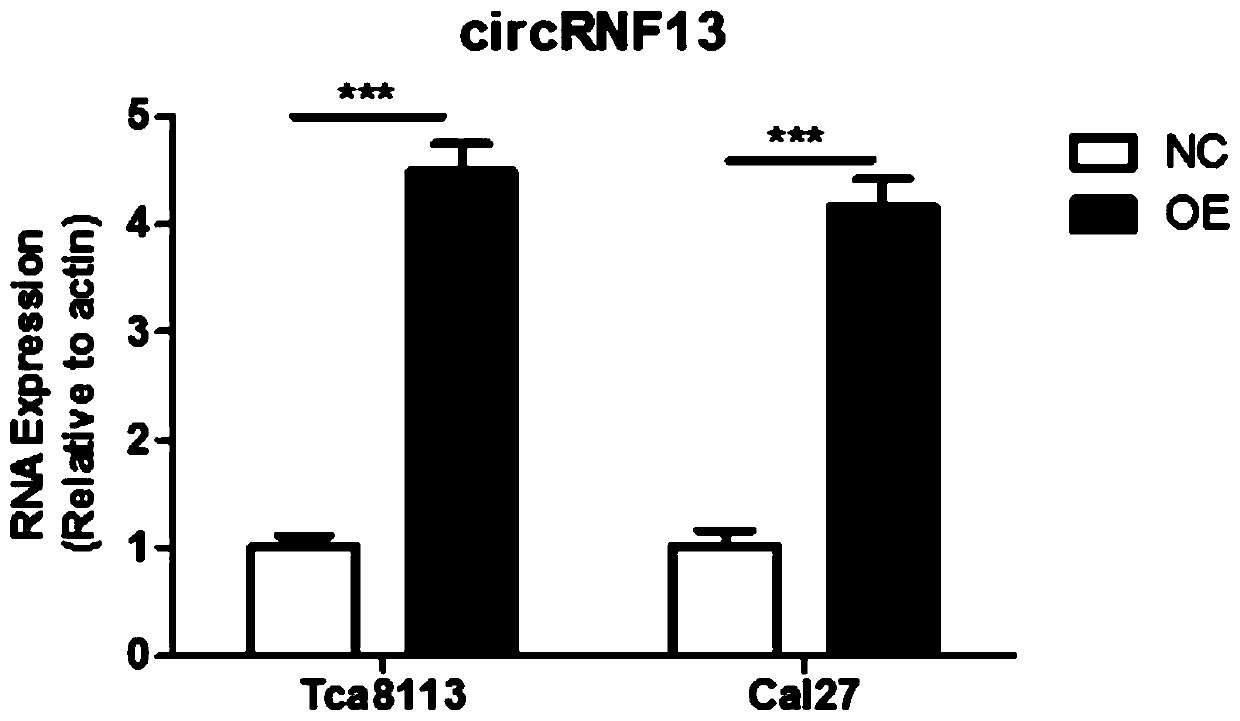
[0084] Example 2, overexpression of circRNF13 in tongue squamous cell carcinoma cells
[0085] 1. Materials and methods
[0086] 1.1 Cell culture and transfection
[0087] The tongue cancer cells Tca8113 and Cal27 in good growth state were divided into 2×10 5 Cells / well were seeded in a 6-well plate, and the 6-well plate was placed at 37°C, 5% CO2 In the incubator, the transfection of the circRNF13 overexpression vector can begin when the cells to be cultured grow to a density of 50-70%; the transfection process is as follows:
[0088] Add 100 μl of the prepared polylysine-modified silicon nanoparticle suspension carrying the circRNF13 eukaryotic expression plasmid to a sterile EP tube, and gently mix with 100 μl of serum-free medium; wash the cells 3 times with D-Hank's solution; Add 800 μl of serum-free medium (without antibiotics) to the above mixture, mix gently and add to one well of a 6-well plate; place the 6-well plate in CO 2 Incubate at 37°C for 6 hours in an incu...
Embodiment 3
[0101] Cell Culture and Transfection
[0102] Tongue squamous cell carcinoma cells Tca8113 and Cal27 in good growth state or drug-resistant cells were divided into 2×10 5 Cells / well were seeded in a 6-well plate, and the 6-well plate was placed at 37°C, 5% CO 2 In the incubator, the transfection of circRNF13 overexpression vector or siRNA can be started when the cultured cells grow to 50-70% density; the transfection process is as follows:
[0103] Add 100 μl of prepared polylysine-modified silicon nanoparticle suspension carrying circRNF13 eukaryotic expression plasmid or siRNA to a sterile EP tube, mix gently with 100 μl serum-free medium; wash cells with D-Hank's solution 3 times; add 800 μl serum-free medium (without antibiotics) to the above mixture, mix gently and add to 1 well of the 6-well plate; place the 6-well plate in CO 2 Incubate at 37°C for 6 hours in an incubator, discard the supernatant, and add complete medium to continue culturing overnight. Silicon nanop...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com