Methods for constructing and predicting leaf trait of woody plant and photosynthetic characteristic model based on DNA methylation level
A technology of leaf phenotype and photosynthetic characteristics, applied in the field of biological information, can solve the problems of huge data volume, high research cost, poor accuracy of results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0200] The following is an example of 235 individuals of Populus microphylla collected across the country.
[0201] Collection of experimental data:
[0202] Using the enzyme fragment A disclosed in Variation in genomic methylation in natural populations of Populus simonii is associated with leaf shape and photosynthetic traits (Journal of Experimental Botany, Vol.67, No.3pp.723-737, 2016) published by Dong Ci et al. The methylation data were calculated to obtain the DNA genome methylation levels of 235 Populus microphylla samples, and the results are shown in Table 1 (CC means Chicheng County: Chicheng County, ZJK: Zhangjiakou FX: Shaanxi Fu County; LY: Linyou County Linyou County; LX: Langao County, LC: Luochuan County, GQ: Gaoling Count; HZ: Huzhu County, XH: Xinghai County; W: DulanCounty, MY: Menyuan County County, SX: Song County, YC: Yichuan County, JL: Zhongning County, NM: Baotou City, NW: Ningwu County, TRT: Taoranting Park). At the same time, a portable laser leaf...
Embodiment 2
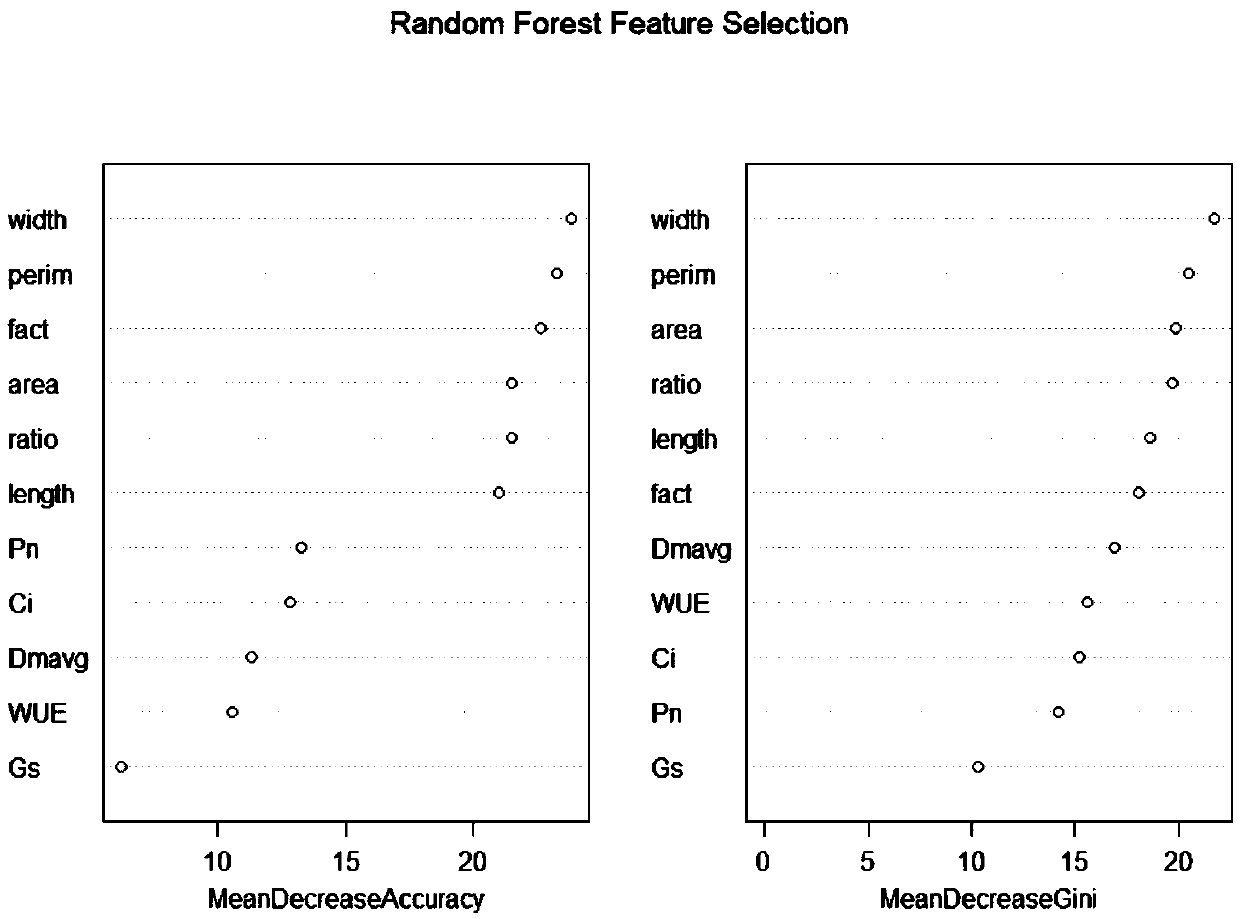
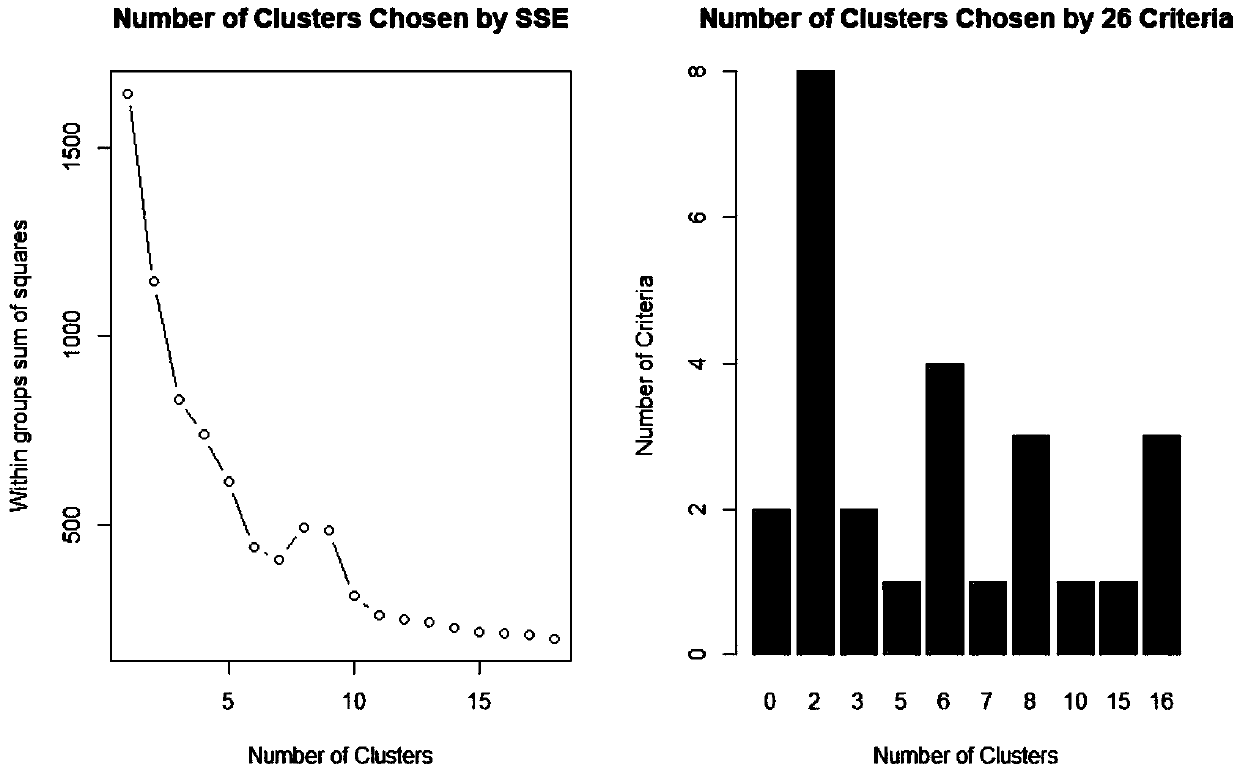
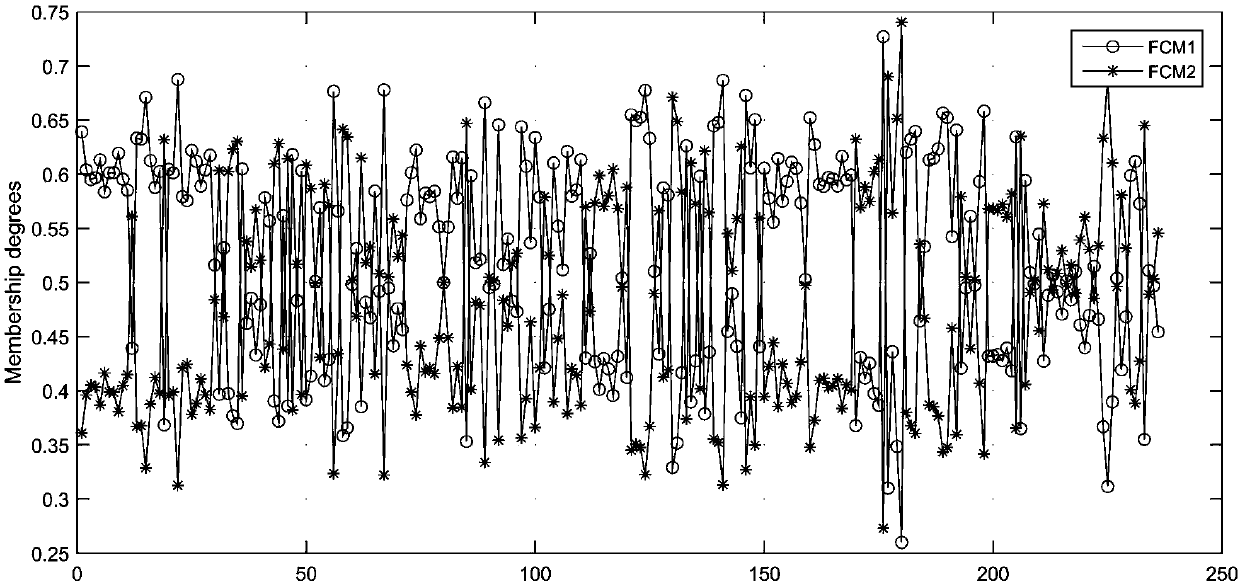
[0217] The relationship between DNA methylation level and leaf phenotype characteristics and photosynthetic characteristics of Populus microphylla.
[0218] To explore the effect of DNA methylation levels on phenotypic traits (leaf shape factor, leaf area, and net photosynthetic rate), we analyzed the numerical values of marginal utility.
[0219] In the first type of poplar samples, the DNA methylation levels of enzyme digestion combinations H31E3, H60E3, H63E10, H44E4, H80E2, H65E5, H63E11 and H60E2 had a significant marginal effect on leaf area (eg Figure 7-1 ). Leaf area decreased with the increase of the DNA methylation level of the restriction enzyme combinations H60E3, H44E4 and H60E2, and increased with the increase of the DNA methylation level of H31E3, H63E10, H80E2, H65E5 and H63E11. In the second type of poplar samples, the marginal effects of the DNA methylation levels of enzyme digestion combinations H80E13, H60E2, H82E5, H60E15, H63E11, H86E7, H80E9, H86E16,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com