Methods for genome assembly, haplotype phasing, and target-independent nucleic acid detection
A genome and nucleic acid technology for genome assembly, haplotype phasing, and target-independent nucleic acid detection, which can solve problems such as difficult data sets
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach 89
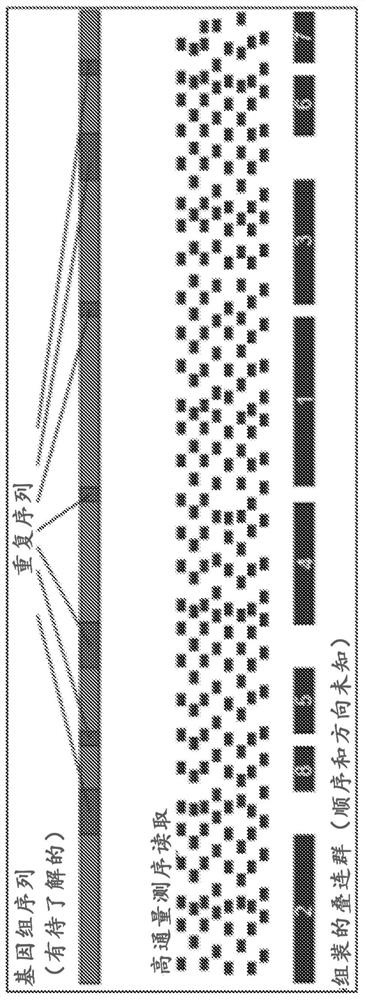
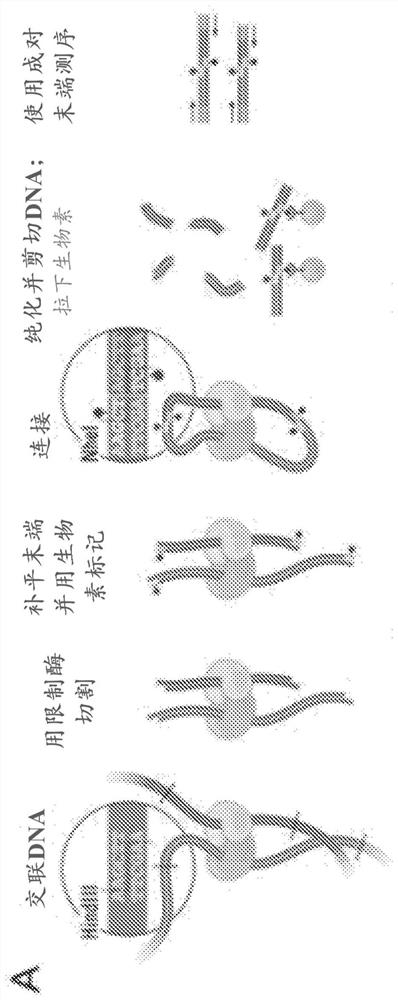
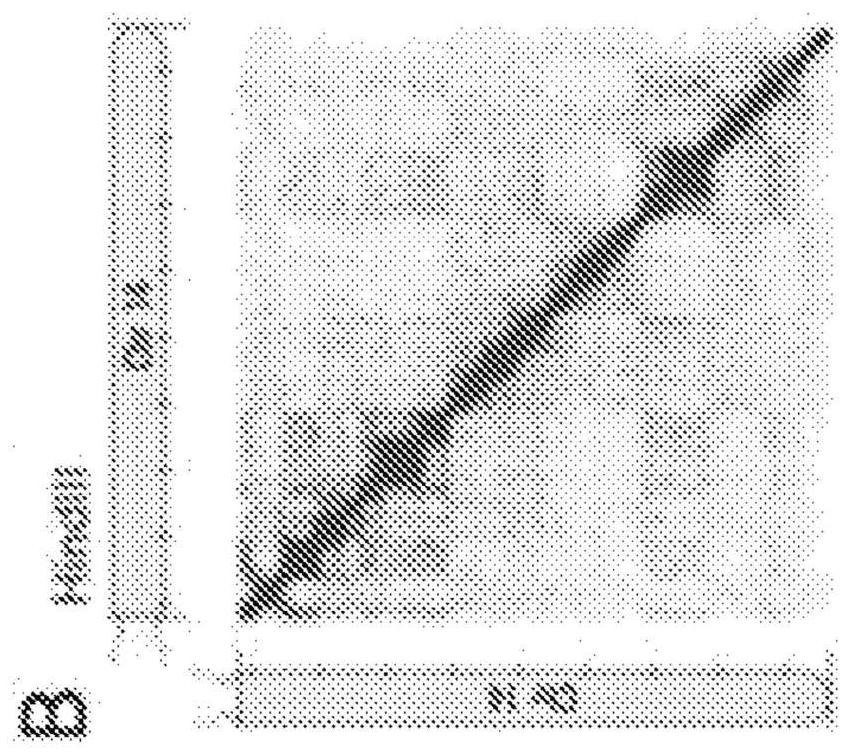
[0431]Numbered embodiment 89 includes a method for generating a plurality of tagged sequences from a plurality of DNA molecules, the method comprising: (a) binding the plurality of DNA molecules to a plurality of association molecules; (b) cleaving the plurality of DNA molecules; DNA molecules to generate a plurality of DNA segments; (c) labeling at least a portion of the DNA segments to form a plurality of marked DNA segments; and (d) sequencing the marked DNA segments to obtain A sequence of a plurality of tags; wherein the plurality of associated molecules are not covalently modified with an affinity tag before or during steps (a) and (b). Numbered embodiment 90 includes the method of numbered embodiment 89, wherein prior to step (b), less than 40% of the DNA segments from the DNA molecule are joined to other DNA segments that do not share a common phosphodiester bond . Numbered embodiment 91 includes the method of any one of numbered embodiments 89-90, wherein prior to st...
Embodiment approach 158
[0433]Numbered embodiment 158 includes a method for generating a plurality of tagged sequences from a plurality of DNA molecules, the method comprising: (a) obtaining a plurality of DNA molecules bound to a plurality of associated molecules; (b) cleaving the DNA molecules to generate at least a plurality of DNA segments; (c) labeling at least a portion of the DNA segments to form a plurality of marked DNA segments; and (d) sequencing the marked DNA segments to obtain a plurality of A labeled sequence; wherein the total amount of said plurality of DNA molecules is less than about 5 micrograms ([mu]g). Numbered embodiment 159 includes a method for generating a plurality of tagged sequences from a plurality of DNA molecules, the method comprising: (a) obtaining a plurality of DNA molecules bound to a plurality of associated molecules; (b) cleaving the DNA molecules to generate at least a plurality of DNA segments; (c) labeling at least a portion of the DNA segments to form a pl...
Embodiment approach 229
[0435] Numbered embodiment 229 includes a method of identifying a microbial host of an antibiotic resistance gene, the method comprising: a) obtaining a stabilized sample from an individual suffering from a condition exhibiting microbial antibiotic resistance; b) processing said stabilized sample to cleavage of double-stranded DNA in the stabilized sample; c) labeling the exposed DNA ends; d) ligating the labeled exposed DNA ends to form labeled paired ends; and e) sequencing the labeled paired ends to Paired sequences are generated; where the sequence adjacent to the antibiotic resistance gene sequence indicates the microbial host of the antibiotic resistance gene. Numbered embodiment 230 includes the method of numbered embodiment 229, wherein the stabilized sample has been crosslinked. Numbered embodiment 231 includes the method of any one of numbered embodiments 229-230, wherein the stabilized sample has been contacted with formaldehyde. Numbered embodiment 232 includes th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com