SgRNA screening for DJ-1 gene editing as well as vector and application of sgRNA
A technology of DJ-1 and genes, applied in fields including sgRNA and its vectors and applications, can solve problems such as time-consuming, laborious, inaccurate, and high requirements for equipment and equipment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
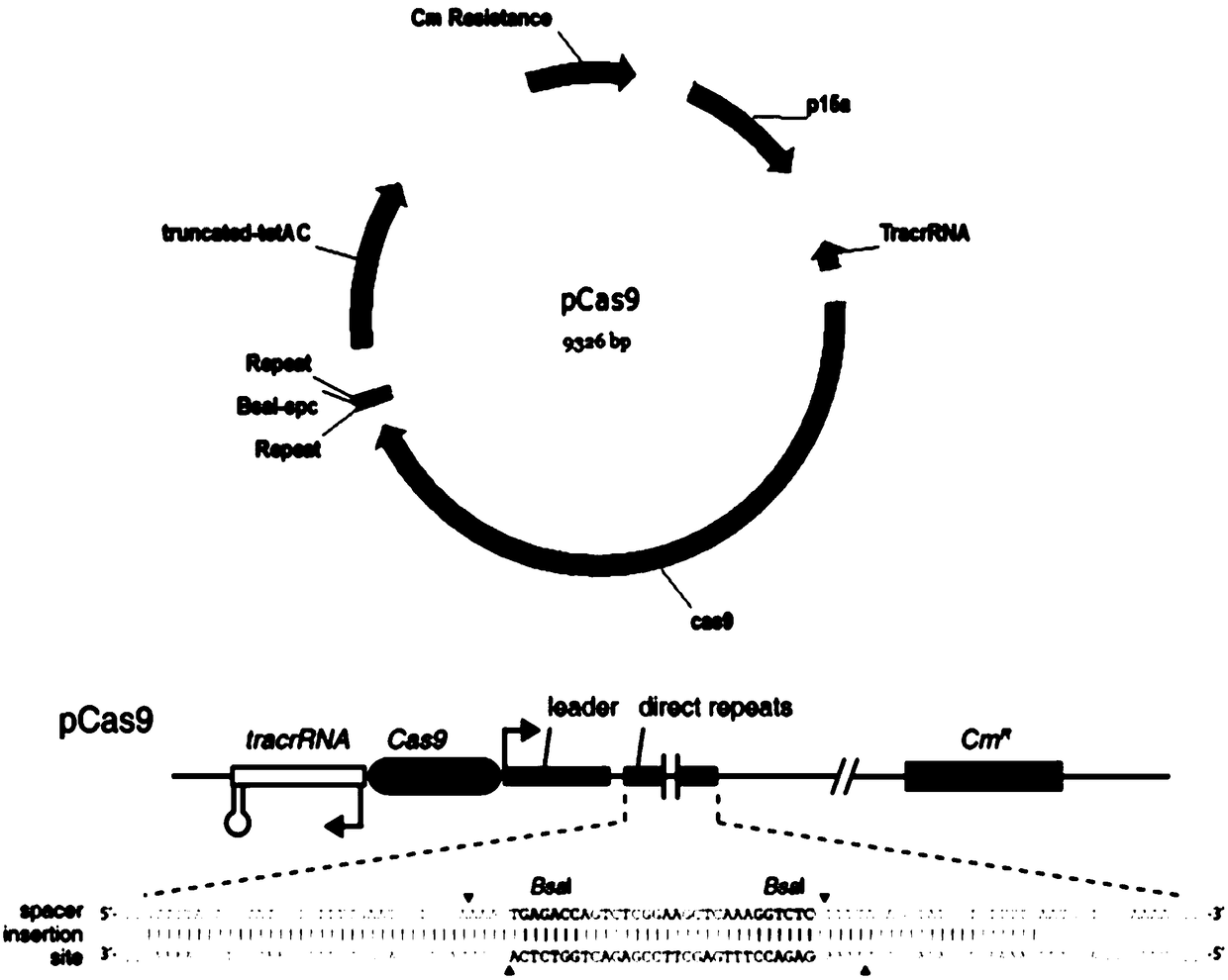
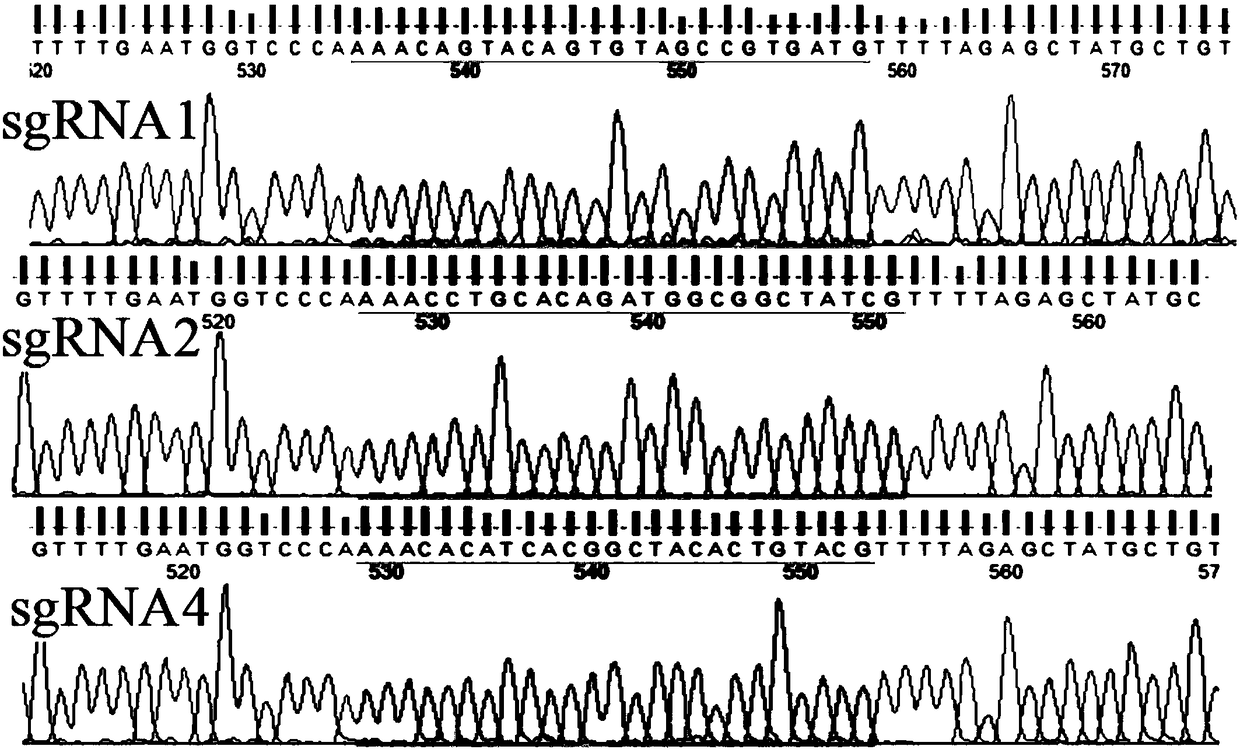
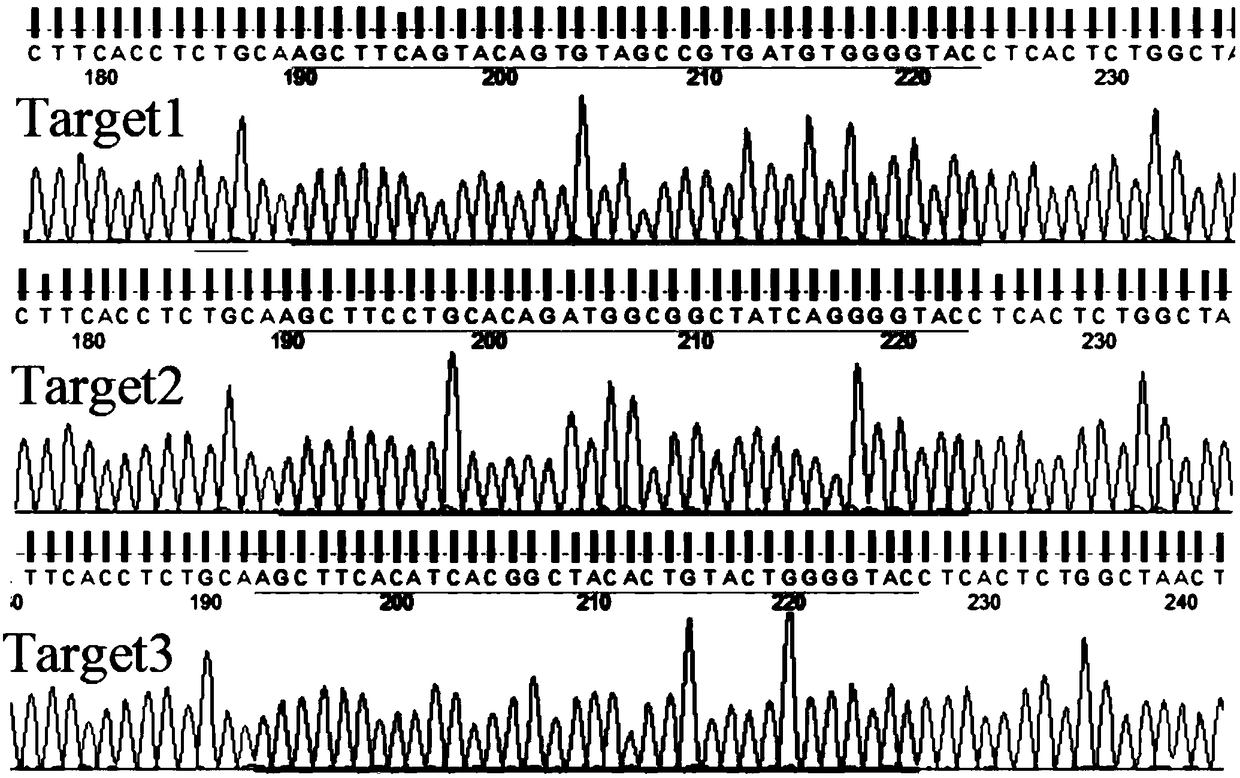
[0058] For the sgRNA of the DJ-1 gene, the sequence of the sgRNA for the DJ-1 gene is SEQ ID NO. 1, specifically 5'-ACATCACGGCTACACTGTACTGG-3', referred to as sgRNA4.
Embodiment 2
[0137] The pSpCas9(BB)-2A-GFP plasmid loaded with the sgRNA (SEQ ID NO. 1) of the present invention was constructed according to the above-mentioned conventional method. According to the above conventional transfection system, pipette 0.8μg of successfully constructed pSpCas9(BB)-2A-GFP plasmid and 2μL of Lipo2000 into centrifuge tubes with 50μL of preheated OMEM medium, mix gently, and let stand for 5min; Then mix the two tubes of solution, shake slightly to mix, and let stand for 20 min. Aspirate the cell culture medium, wash with PBS solution, add 100μL of mixed solution and 100μL of preheated OMEM medium to each well of the 24-well plate, and put in 5% CO 2 Incubate at 37℃ for 4h, then add 200μL of complete medium, incubate for 12h, suck off the liquid in the culture plate, add 1mL of complete medium, and put in 5% CO 2 Incubate at 37°C for 48h. At the same time, the experimental group (containing the sgRNA of SEQ ID NO.1) and the negative control group (without sgRNA) were...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com