Application of long-chain non-coding RNA DUXAP8 as biomarker for prognosis of bladder cancer
A long-chain non-coding, biomarker technology, applied in the field of biomarker application, can solve the problems of unclear clinical significance and potential function of lncRNAs, and low patient survival rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
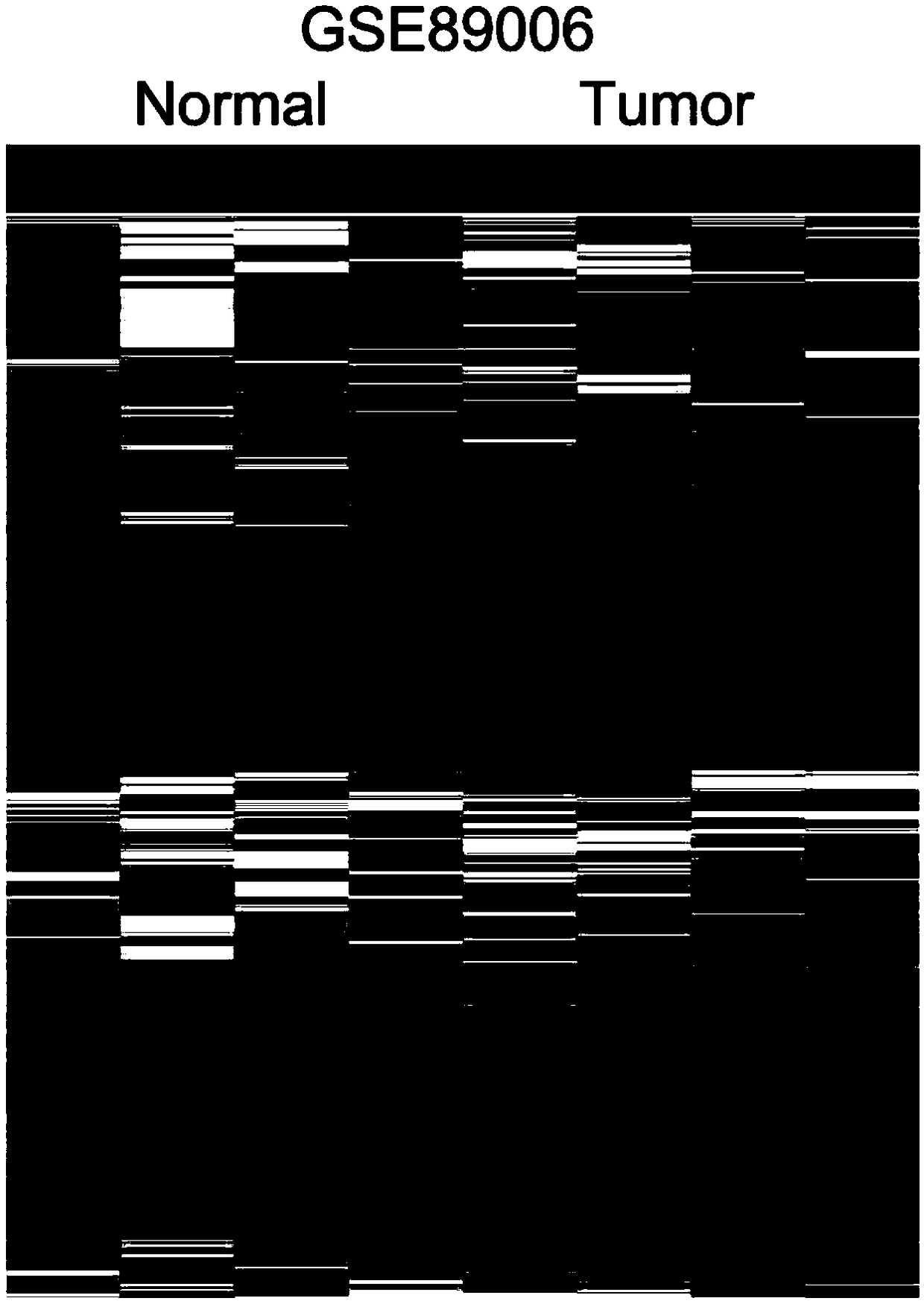
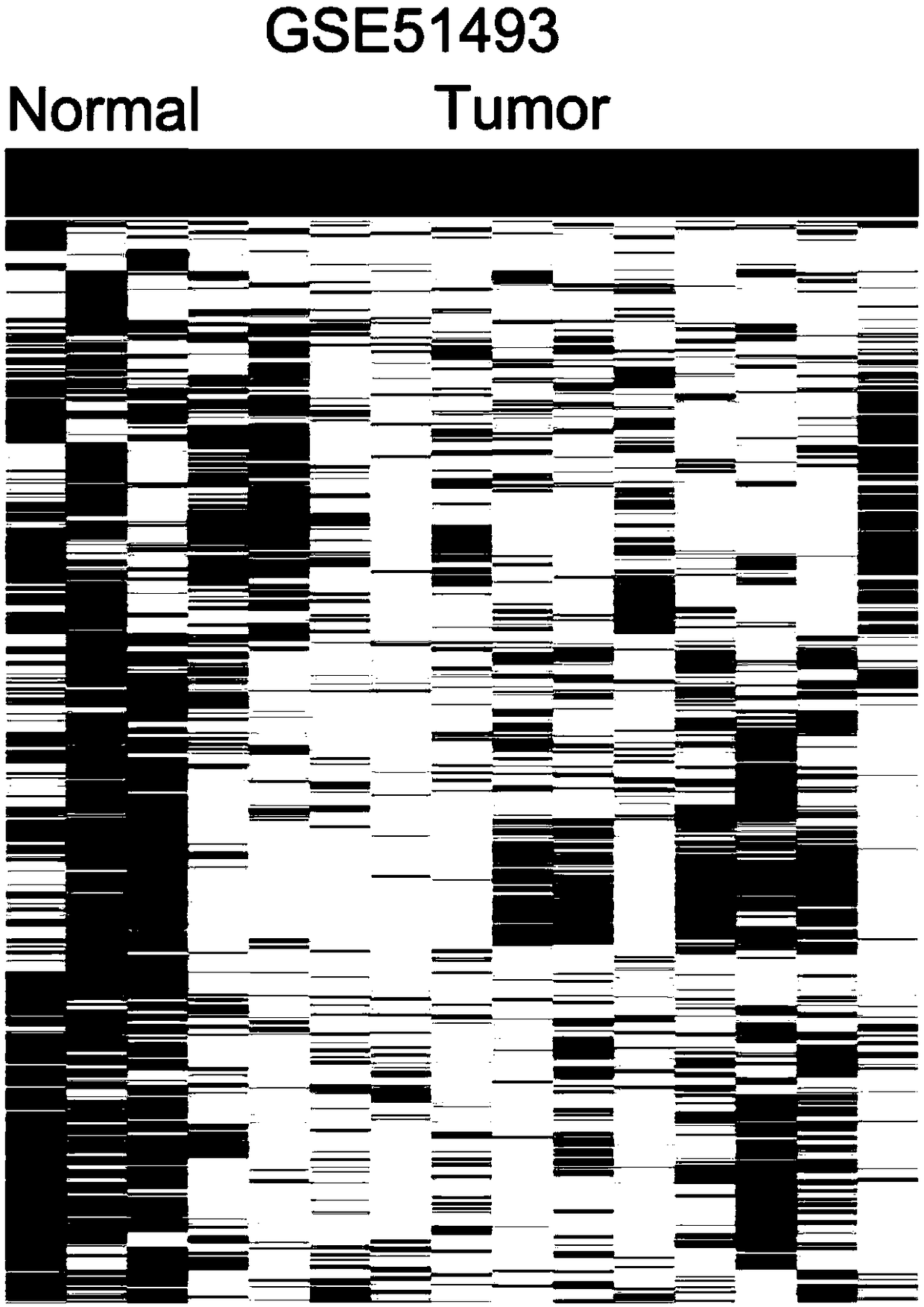
[0036] lncRNA profiling in TCGA and public microarray data
[0037] TCGA bladder cancer tissue and non-tumor tissue RNA-seq data and corresponding clinical information were downloaded from http: / / ibl.mdanderson.org / tanric / _design / basic / download.html. Three other published bladder cancer gene microarray analysis data (GSE89006, GSE51493 and GSE45184) were downloaded from Gene Expression Omnibus (GEO). GSE89006, GSE51493 and GSE45184 were analyzed using Agilent-045997 Arraystarhuman lncRNA microarray V3, ArraystarHuman LncRNA microarray V2.0, and Agilent-028004 SurePrint G3 Human GE 8x60K microarray platforms. These TCGA RNA-seq data and microarray data were preprocessed using R software and software packages.
[0038] Copy number variation analysis of lncRNA loci
[0039] Raw bladder cancer tissue gene copy number variation data were downloaded from the Broad GDAC FireBrowser website. GISTIC 2.0 was used to identify significant recurrent occurrences of copy number deletions or...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com