Genome assembly method for constructing overlength continuous DNA (DeoxyriboNucleic Acid) sequence
A DNA sequence and genome assembly technology, applied in the field of genome assembly, can solve the problems of unable to truly restore the original genome sequence, indistinguishable, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
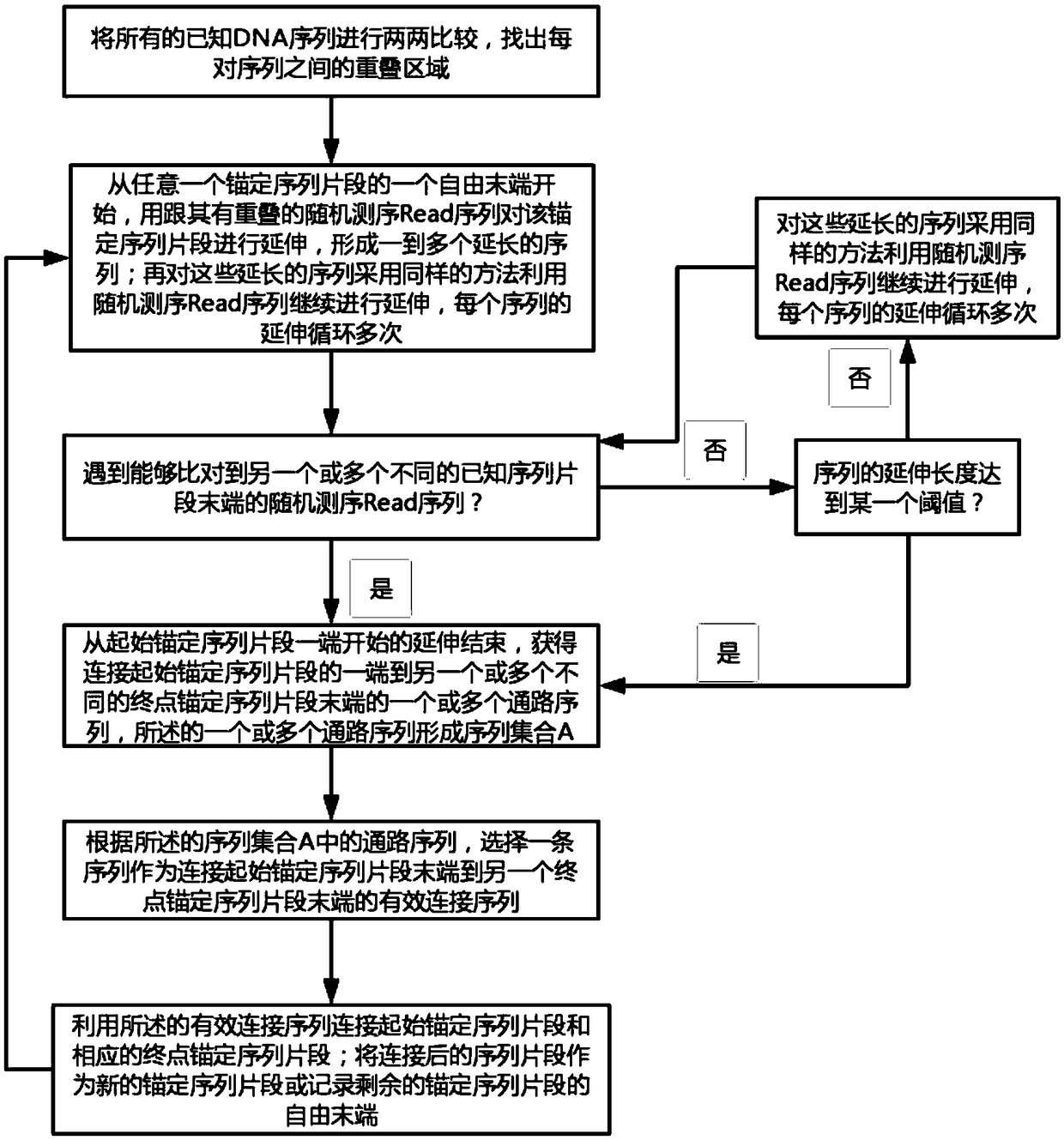
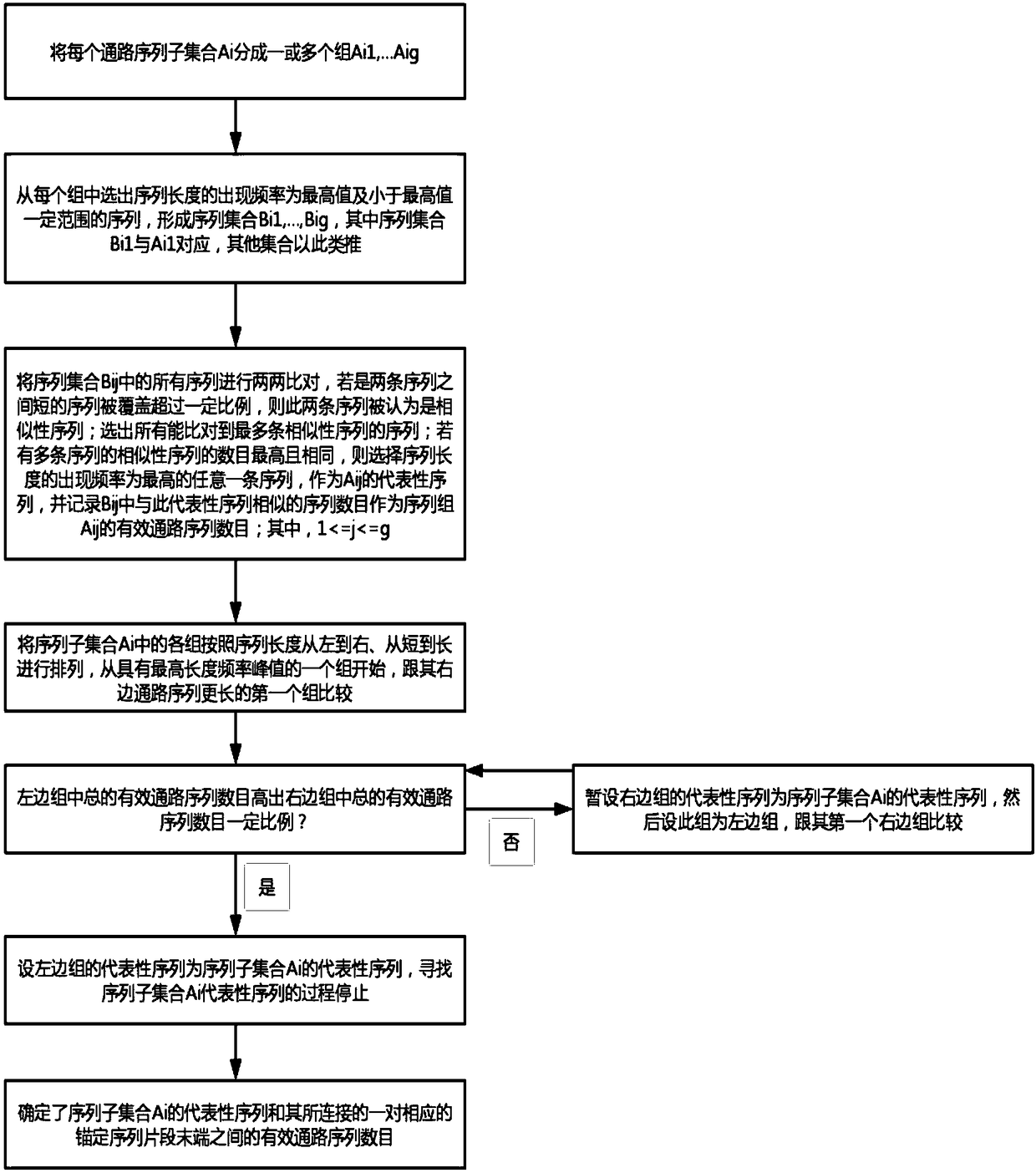
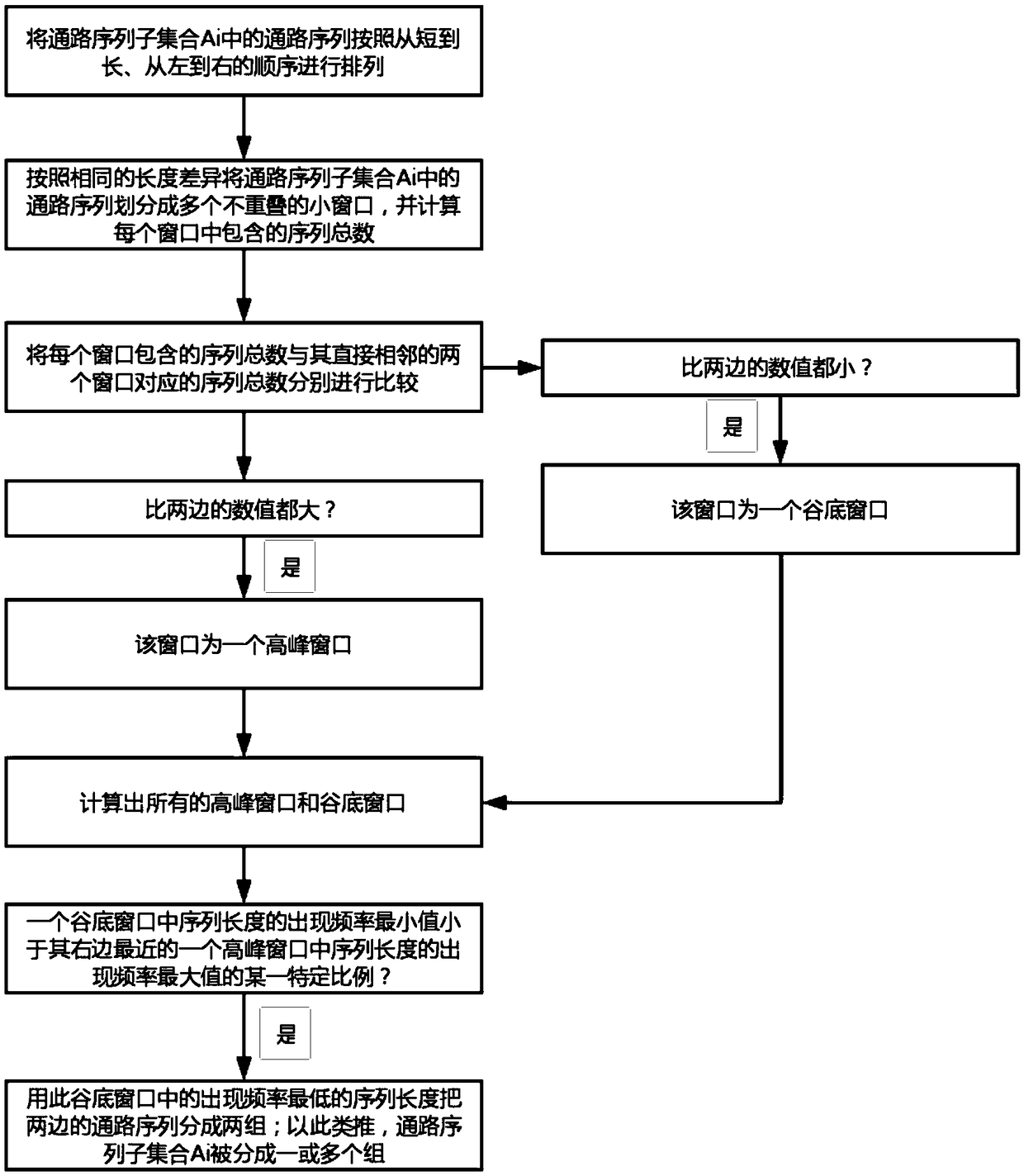
[0064] Embodiment of the present invention: a genome assembly method for constructing ultra-long continuous DNA sequences, such as figure 1 shown, including the following steps:
[0065] S1, compare all known DNA sequences pairwise, and find similar overlapping regions between each pair of sequences; wherein, the known DNA sequences include anchor sequence fragments (that is, sequence fragments used for anchoring, For example, one or several specific sequence fragments intercepted from the DNA sequence, and / or one or several specific sequence fragments that have been assembled, and / or one or several specific sequence fragments selected from random sequencing Read sequences A specific Read sequence, etc.) and random sequencing Read sequence (in order to improve the accuracy of the Read sequence, the Read sequence can be corrected first, or the original random sequencing Read sequence can be directly used without correction; the correction method includes using The sequencing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com