Genes for molecular typing of hypermutated tumors and their application
A mutant and tumor technology, applied in the determination/testing of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the defects of gene mismatch repair system, microsatellite instability, and can not well prompt immunity Issues such as the response to treatment and the possibility of ultimate benefit to achieve good benefits and good clinical guidance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Preparation of Genomic DNA Samples and Determination of Tumor Somatic Mutation Sites
[0031] In order to detect the somatic mutation of colorectal cancer, the present invention completed the high-throughput sequencing of 338 colorectal cancer tissue samples in two stages, and completed the high-throughput sequencing of 80 cases of colorectal cancer in the first stage, of which 10 cases For whole genome sequencing, 70 cases for whole exome sequencing. Through the first stage of high-frequency gene analysis, further combining the high-frequency genes in the TCGA database and COSMIC database, and the NCCN guidelines for the diagnosis and treatment of hereditary colorectal cancer, the present invention designed a gene group comprising 524 genes (Table 1), Capture probes targeting this gene group were customized for the second stage of sequencing. In the second phase, the targeted sequencing of 258 colorectal cancer tissue samples was completed using the capture ...
Embodiment 2
[0034] Example 2 Establishment of a gene mutation prognosis prediction model for hypermutated colorectal cancer
[0035] Since the currently approved immunotherapy drugs are mainly aimed at tumor patients with dMMR and MSI‐H, the characteristics of these patients at the genomic level are mainly hypermutations. Tumors with a mutation burden greater than 10 Mut / Mb were defined as hypermutated tumors. Among the 338 cases of colorectal cancer (ZJU data set) in Example 1, 45 cases were determined to be hypermutated. In addition, in order to verify the stability and universality of the model, the present invention downloaded a total of 382 cases of colorectal cancer data from TGCA as independent verification data (TCGA data set), and according to the same standard, 63 cases were designated as hypermutated colorectal cancer. Further, the present invention requires patient data with a follow-up time of more than 24 months for the establishment of a prognostic model, so 45 cases from...
Embodiment 3
[0037] Example 3 Analysis of colorectal cancer from ZJU and TCGA datasets using DOCK2 gene prognostic prediction model
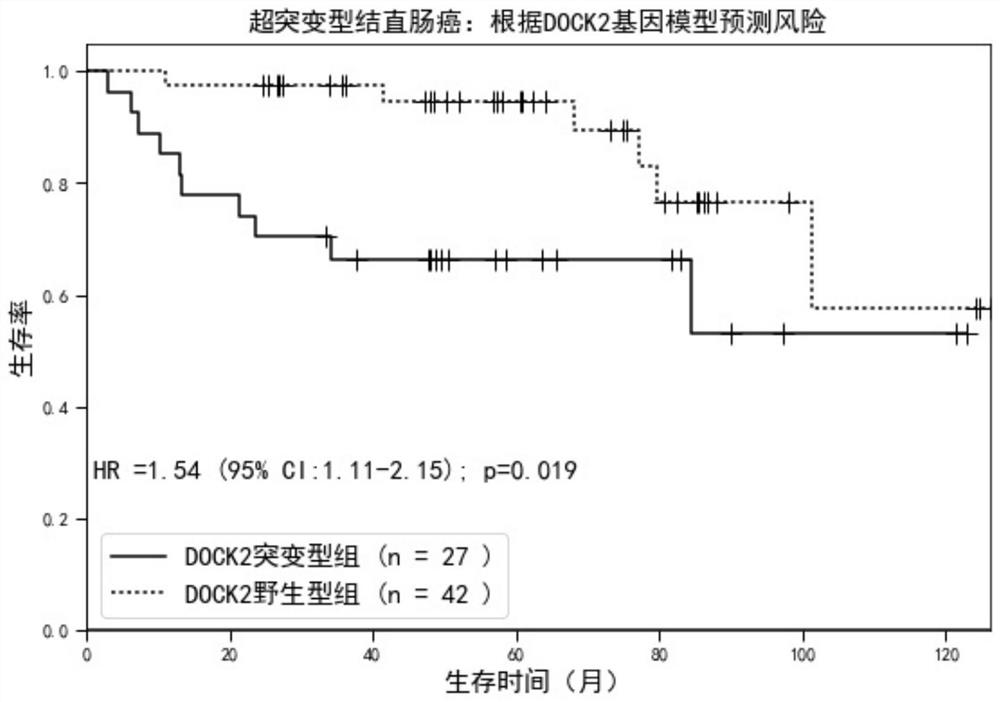
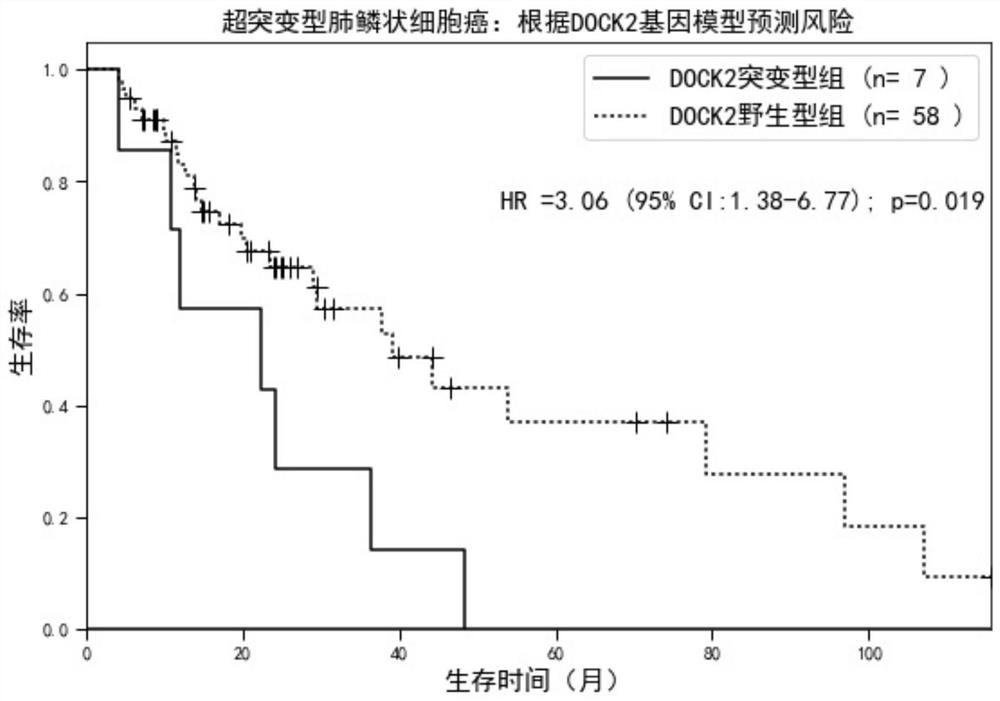
[0038] Since hypermutated and microsatellite unstable colorectal cancers account for about 15% of the total, the number of patients in a single data set is relatively small. In order to further fully demonstrate the beneficial effects of the present invention, the present invention will use the colorectal Integrate the cancer data set, apply the DOCK2 gene prognosis prediction model constructed in Example 2, analyze the hypermutated colorectal cancer in the integrated data set, and distinguish it as DOCK2 wild type according to the mutation status of the DOCK2 gene in each patient in Table 2 group and DOCK2 mutant group, see figure 1 , compared with patients in the DOCK2 wild-type group, the hazard ratio of the DOCK2 mutant group was 1.54 (95% confidence interval = 1.11-2.15, P = 0.019).
[0039] Table 2
[0040]
[0041]
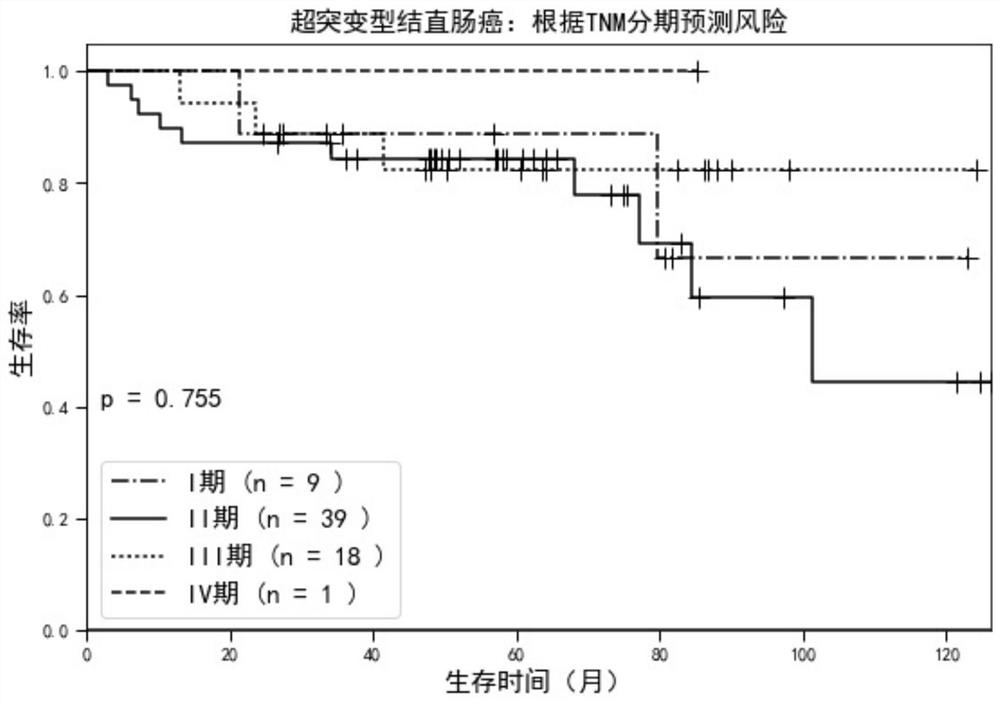
[0042] The TNM staging sta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com