Four SNP site combinations for identifying Tan sheep and non-Tan sheep and application thereof
A non-tan sheep and locus technology, applied in the field of identifying the combination of 4 SNP loci of Tan sheep and non-tan sheep, can solve problems such as Tan sheep brand damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1. Discovery of SNP site combinations for identification of Tan sheep and non-Tan sheep and establishment of detection methods
[0064] 1. Discovery of SNP loci for identification of Tan sheep and non-Tan sheep
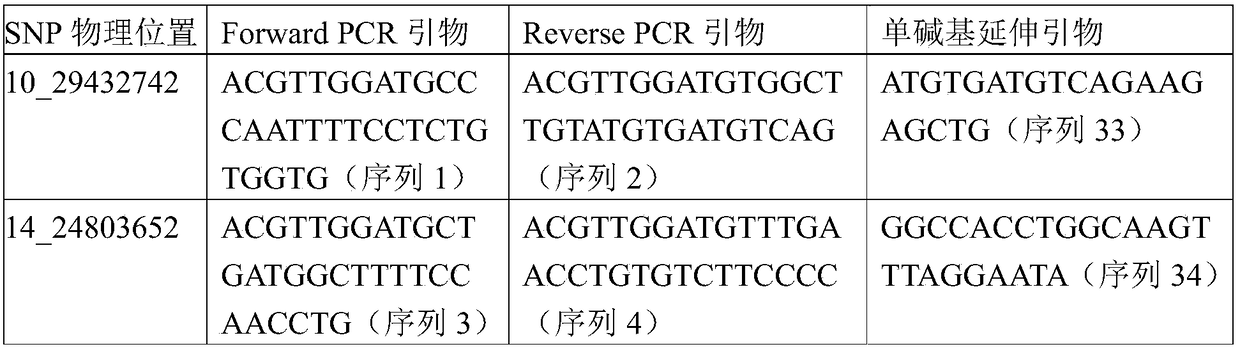
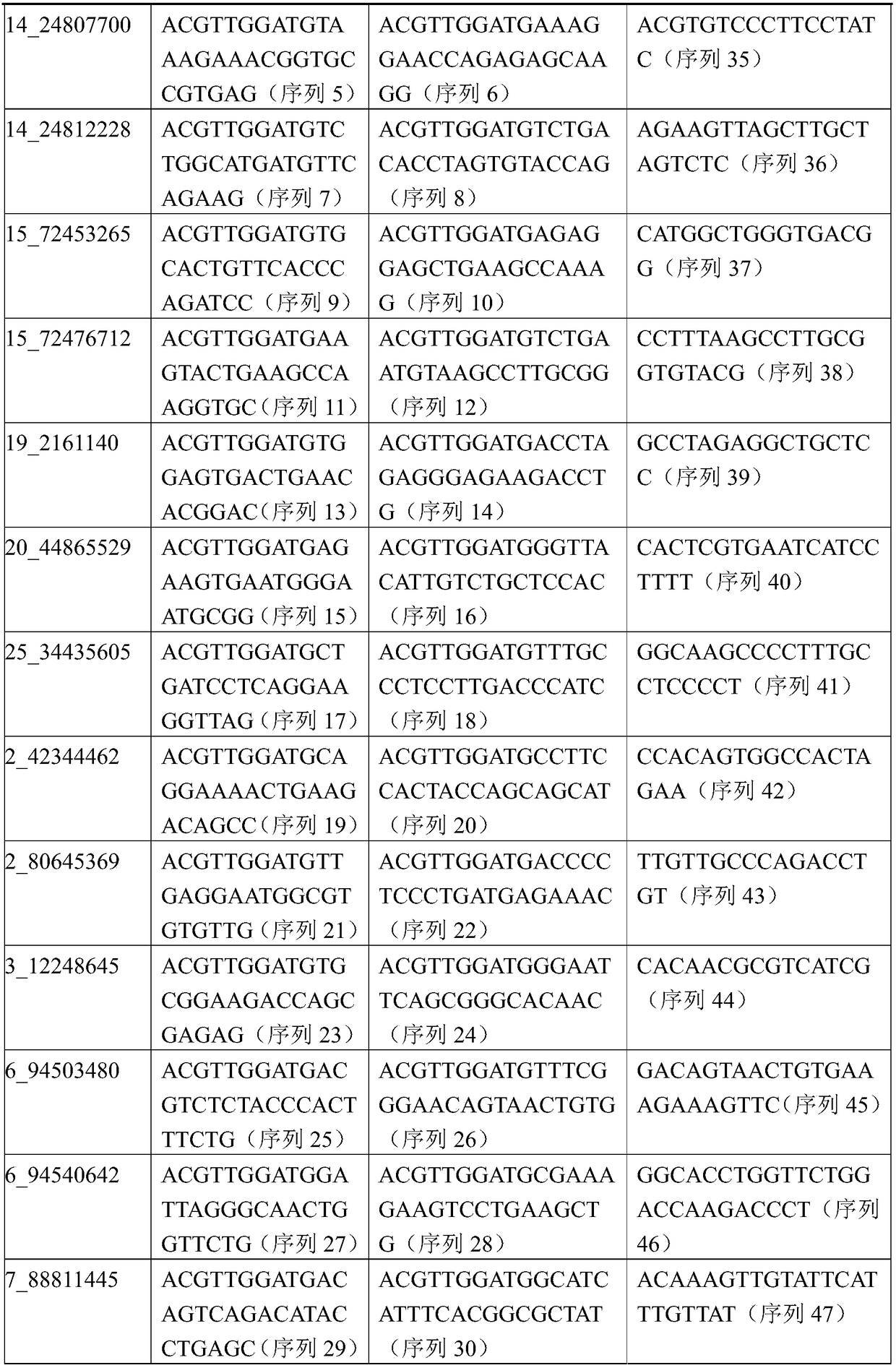
[0065] Genomic DNA was extracted from the blood or ear tissues of 50 Tan sheep (from Yanchi County, Ningxia) and 50 Feitan sheep (from Ningxia, Inner Mongolia, etc.). Genomic DNA was resequenced with the Illumina Hiseq X10 instrument, and then analyzed by R software to obtain 25 SNP sites that are different from Tan sheep and non-Tan sheep, and R programming was used to analyze and delete the sites with poor call rate. 模型分析的16个位点如下:10_29432742、14_24803652、14_24807700、14_24812228、15_72453265、15_72476712、19_2161140、20_44865529、25_34435605、2_42344462、2_80645369、3_12248645、6_94503480、6_94540642、7_88811445、7_89505361。
[0066] The physical positions of the 16 SNP sites were determined based on the comparison of the standard sequence of the whole genome of Ta...
Embodiment 2
[0178] Example 2. Blind test-verification of the accuracy of identifying Tan sheep and non-Tan sheep by combining four SNP loci
[0179] Sample: 576 Tan sheep and non-Tan sheep
[0180] Experimental method: use the method for identifying or distinguishing Tan sheep and non-Tan sheep established in Example 1 to detect the genotype of each sample, and obtain the genotyping information of the 4 SNP site combinations of each individual. Compare the identification results with the real results given by experts to test the identification accuracy, false positive rate and false negative rate. Based on the results of the first blind test, the model is optimized by optimizing the training set through machine learning. According to the optimized model, the samples were tested again blindly, and the accuracy rate, false positive rate and false negative rate were evaluated.
[0181] Experimental results: A total of 419 Tan sheep samples and 151 non-Tan sheep samples were identified in t...
Embodiment 3
[0182] Example 3. Blind Test II Verifies the Accuracy Rate of Identification of Tan Sheep and Non-Tan Sheep by Combination of Four SNP Loci
[0183] Sample: 768 Tan sheep and non-Tan sheep
[0184]Experimental method: use blind test-optimized training set to detect the genotype of each sample by using the method of identifying or distinguishing Tan sheep and non-Tan sheep constructed in Example 1 for 768 samples, and obtain 4 kinds of SNP positions of each individual Point combined genotyping information, conduct a second blind test, compare the identification results with the given results, and calculate the identification accuracy rate, false positive rate and false negative rate.
[0185] Experimental results: 679 of the 768 samples were genotyped, and a total of 503 Tan sheep samples and 176 non-Tan sheep samples were identified by the blind test. Compared with the actual species of the given samples, the identification accuracy rate was calculated to be 68.5 %, the false...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com