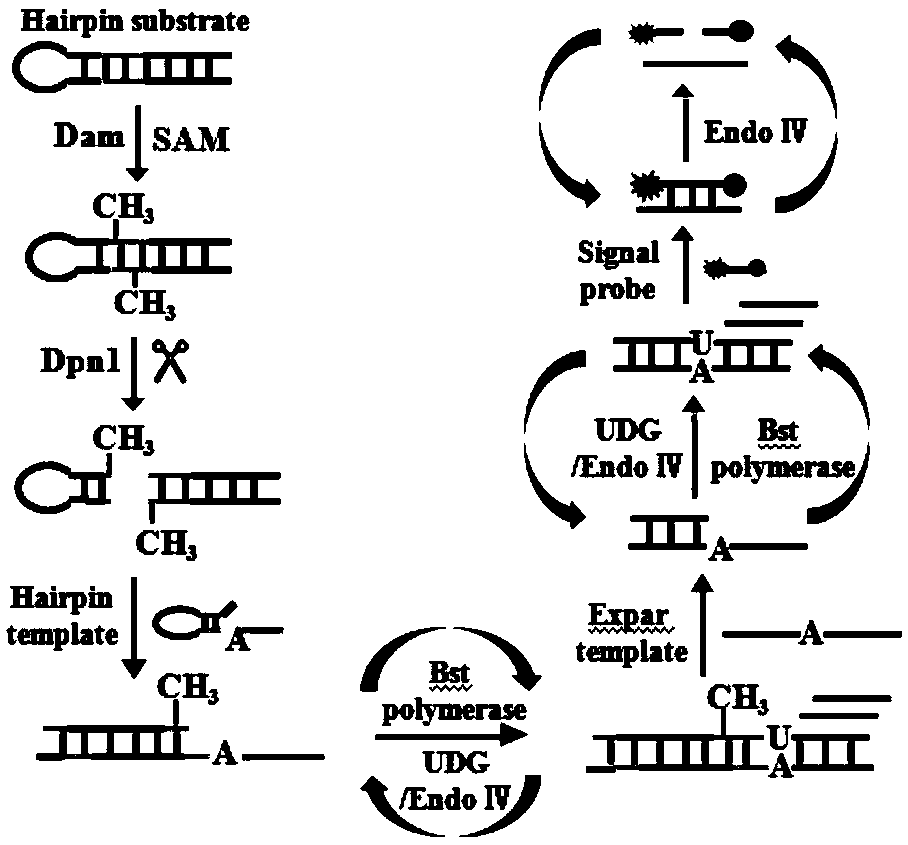
Method for detecting Dam methyltransferase activity based on base excision repair induction
A methyltransferase and detection method technology, applied in the field of biological detection, can solve the problems of reducing the sensitivity of the analysis method, cumbersome operation steps, long analysis time, etc., and achieve the effects of improving the detection sensitivity, good selectivity, and highly sensitive detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 A method for detecting Dam methyltransferase activity based on base excision repair-induced strand displacement and isothermal exponential amplification fluorescence
[0034] Dam methyltransferase activity assay:
[0035] All nucleic acids were diluted to 100 μM with EDTA buffer before use, then the EXPAR template and signal probe were diluted to 1 μM and 10 μM with water, respectively, and the hairpin substrate and hairpin template were annealed in a 95°C water bath for 5 min, and then naturally cooled to room temperature so that the final concentrations were 5 μM and 1 μM, respectively.
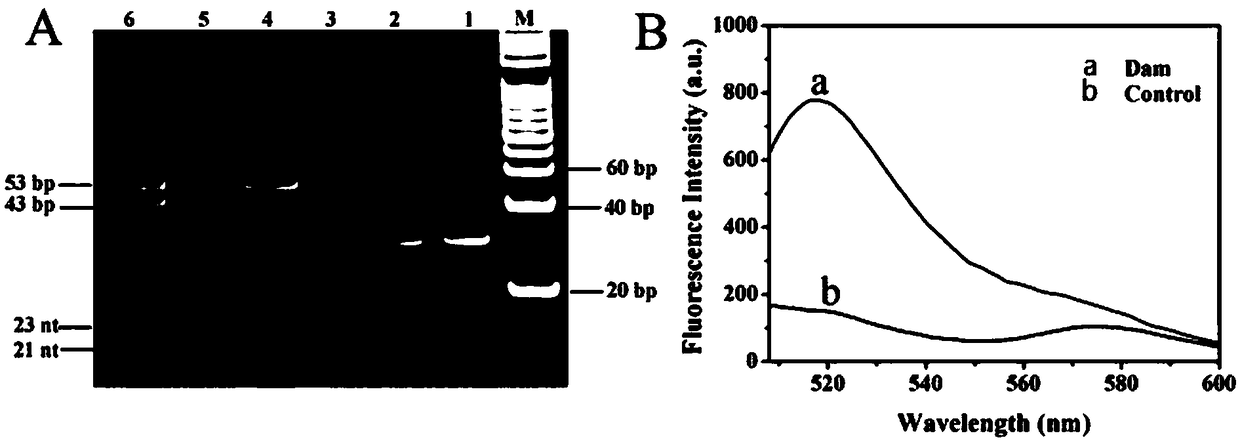
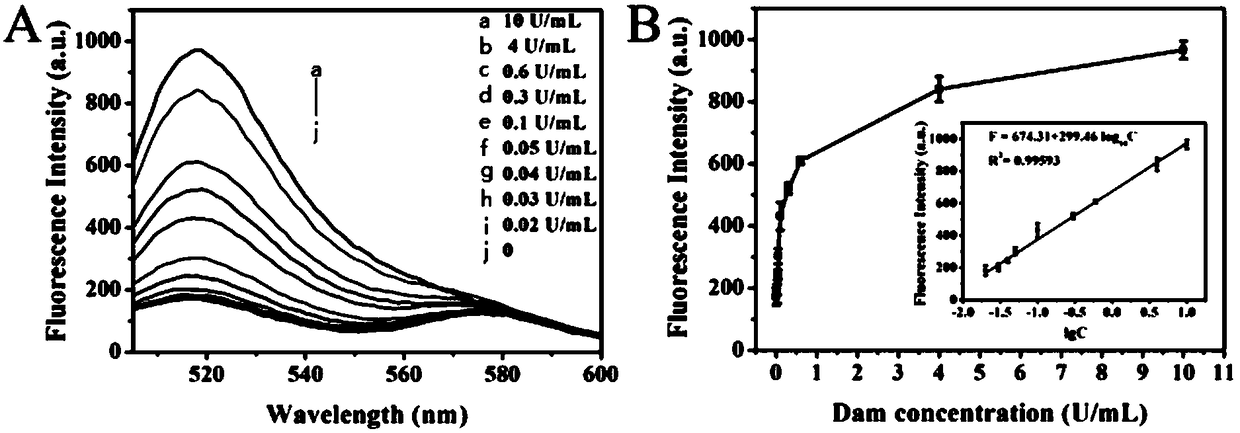
[0036] Add 0.5 μM hairpin substrate, 1×dam buffer, Buffer, 160μM SAM, 10U Dpn I and different concentrations of Dam were reacted at 37°C for 2h and then inactivated at 80°C for 20min. Then, in a 20 μL reaction system, add 4 μL of the above methylated cleavage product, 50 nM hairpin template, 100 nM EXPAR template, 350 nM signal probe, 2.8U Bst DNA polymerase, 1U UDG, 5U ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com