Method for preparing core-shell SERS structure based on amplification of nucleic acid strand by terminal deoxynucleotidyl transferase
A technology for terminal transferase and nucleic acid amplification, which is applied in the field of preparation of nucleocapsid SERS structures, can solve the problems of few and rare preparations of nucleocapsid SERS structures based on DNA bases, and achieve high-efficiency and uniform SERS effects, simple methods, and The effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
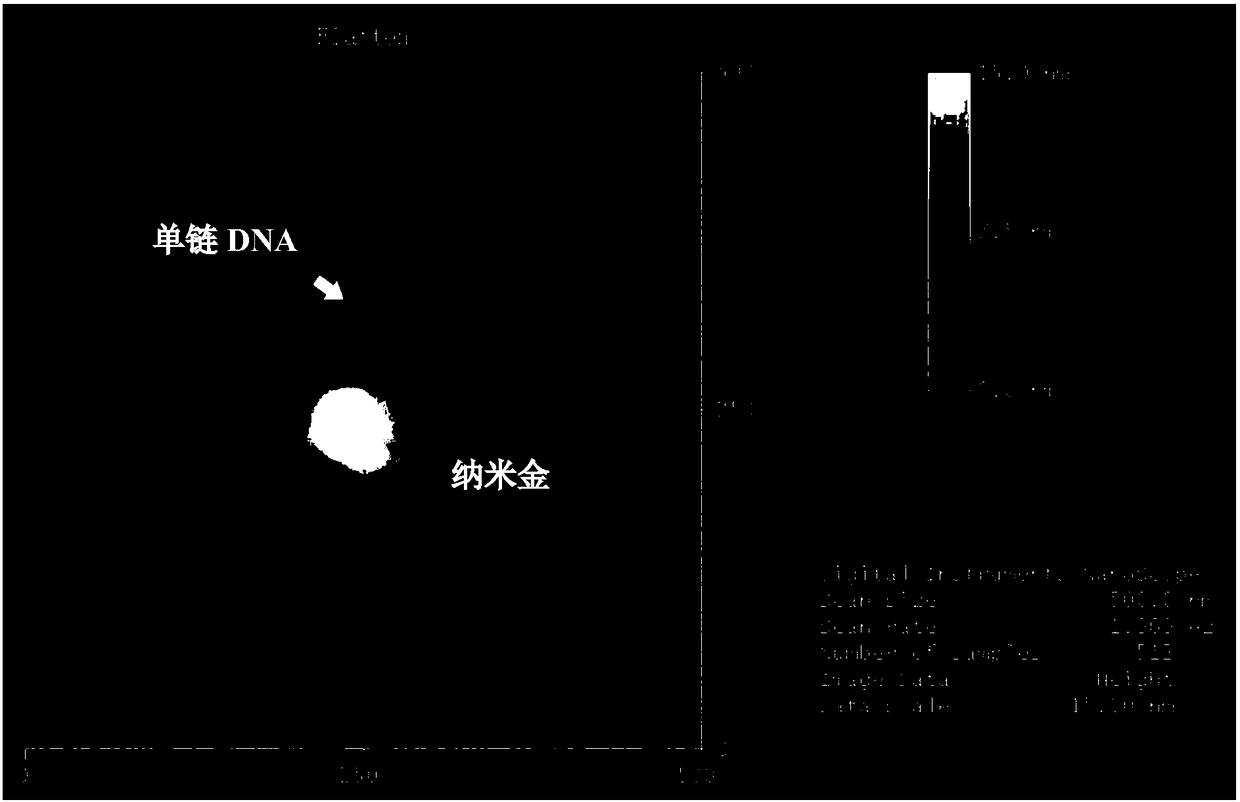
[0036] (1) Add 10 μL thiol-modified initial primer chain (SH-polyA 10 , 100 μM), diluted with water to the final concentration of the initial chain is 5 μM. After shaking at room temperature overnight, add 100mM PB buffer (phosphate buffer) until the final concentration of phosphate buffer is 10mM; after overnight at room temperature, add aging solution (10mM PB, 2M NaCl, pH7.4) so that the final concentration of NaCl in the solution is 0.15 M.
[0037] After the mixture solution was left overnight at room temperature, it was centrifuged at 4°C and 12,000 rpm for 20 min, and washed by centrifugation for 4 times. The precipitate, namely the nanogold-starting chain complex, was resuspended in 200 μL of PB (10 mM, 0.15 M NaCl, pH 7.4) solution, Store at 4°C.
[0038] (2) 4 μL nano gold-starting chain complex (ie Au-PolyA 10 , the 5'-end is connected to the surface of gold nanoparticles), add 1 μL terminal transferase (concentration 10U / μL), 1 μL dATP (concentration 10mM), 2 ...
Embodiment 2
[0045] (1) 10 μL of thiol-modified initial chain (SH-polyT 15 , 100 μM), the starting chain final concentration was 5 μM. After shaking at room temperature overnight, add 100mM PB buffer (phosphate buffer) until the final concentration of phosphate buffer is 10mM. After overnight at room temperature, add aging solution (10mM PB, 2M NaCl, pH 7.4) to make the final concentration of salt in the solution 0.15M . After overnight at room temperature, the mixture solution was centrifuged and washed 4 times at 4°C at 12000rpm / min for 20min, and the precipitate was resuspended in 200μL of PB (10mM, 0.15M NaCl, pH7.4) solution and stored at 4°C.
[0046] (2) 4 μL complex gold-starting chain (Au-PolyT 15 ), add 1 μL terminal transferase (concentration 10U / μL), 1 μL dTTP (10 mM), 2 μL Reaction Buffer to make up the volume to 10 μL, mix the solution well and incubate at room temperature for 1 h; bathe in 70 ° C for 15 min to inactivate the enzyme .
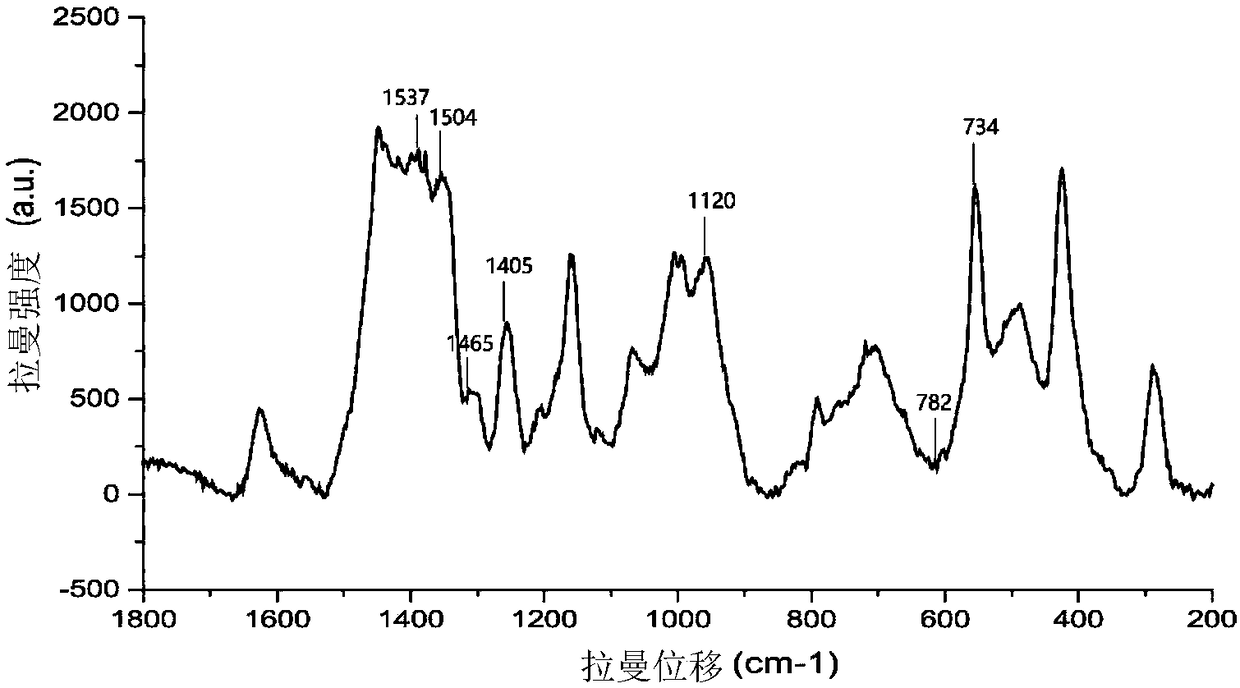
[0047] Take 4 μL of the sample, and...
Embodiment 3
[0050] (1) 10 μL of thiol-modified initial chain (SH-polyA 10 , 100 μM), the starting chain final concentration was 5 μM. After shaking at room temperature overnight, add 100mM PB buffer (phosphate buffer) until the final concentration of phosphate buffer is 10mM. After overnight at room temperature, add aging solution (10mM PB, 2M NaCl, pH 7.4) to make the final concentration of salt in the solution 0.15M . After overnight at room temperature, the mixture solution was centrifuged and washed 4 times at 4°C at 12000rpm / min for 20min, and the precipitate was resuspended in 200μL of PB (10mM, 0.15M NaCl, pH7.4) solution and stored at 4°C.
[0051] Follow the same system and method to assemble the initial chain SH-polyT on the surface of gold nanoparticles 15 , operate with embodiment 2.
[0052] (2) 4 μL complex Au-polyA 10 Add 1 μL of terminal transferase, 1 μL of ddATP (10 mM), 2 μL of Reaction Buffer, mix the solution with a total volume of 10 μL and incubate overnight at ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
| Particle size | aaaaa | aaaaa |
| Thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com