Perl language based automation analysis method for population-specific SNP loci
An automatic analysis and population technology, applied in the fields of genomics, instrumentation, proteomics, etc., can solve the problem of not identifying population-specific SNPs, etc., and achieve the effect of improving convenience, data processing efficiency and server usage efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
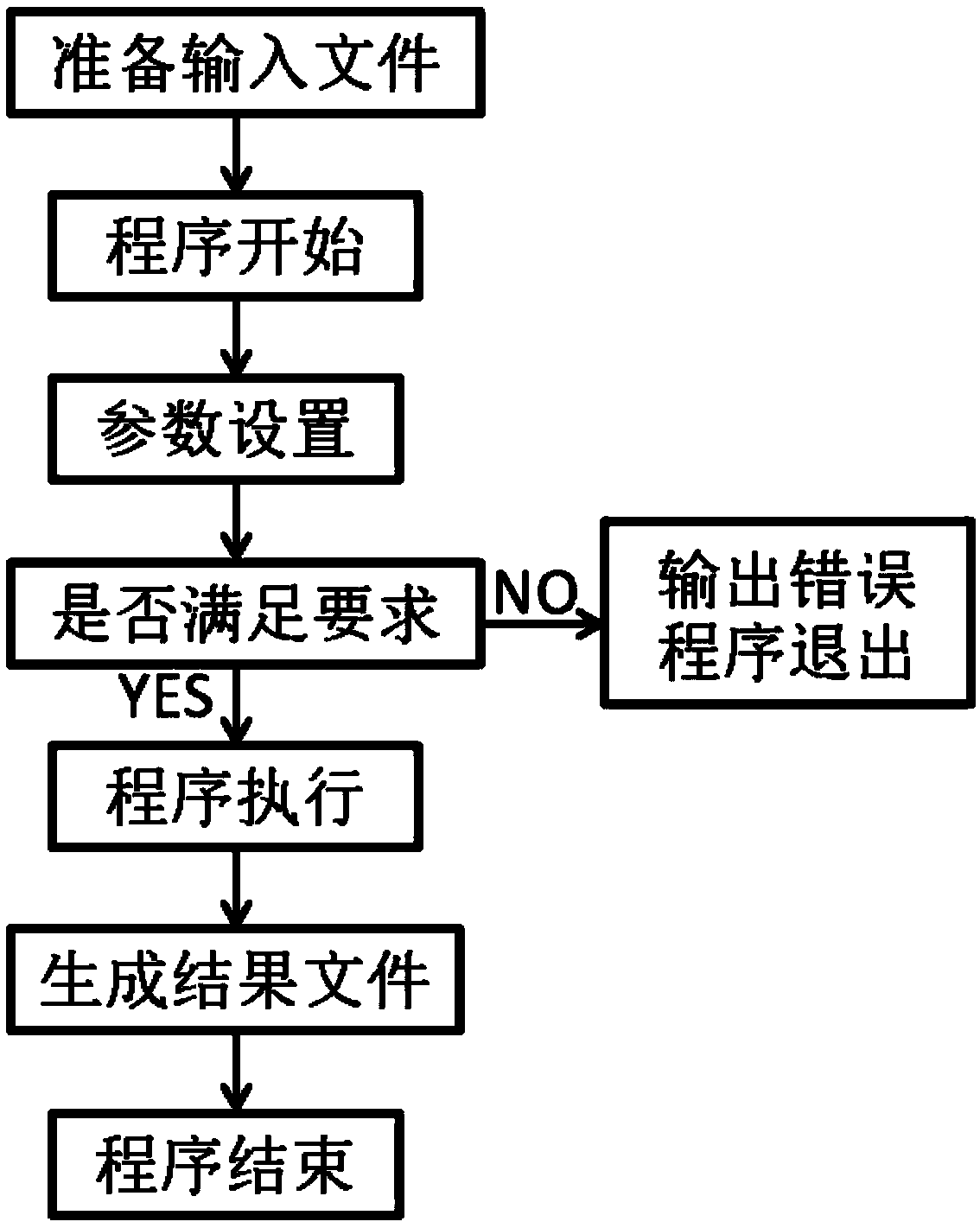
[0022] see figure 1 , a kind of automatic analysis method based on the population-specific SNP site of perl language shown in the figure, comprises the following steps:
[0023] (1) Prepare raw data for analysis;
[0024] The data is a vcf file, which is a common file format for storing variation site information in bioinformatics analysis. Compatible with the result files of all mainstream software for identifying variant sites (samtools / GATK, etc.).
[0025] (2) Set the criteria for SNP site filtering;
[0026] mainly:
[0027] a. The accuracy of the SNP site, that is, the GQ value in the vcf file, the default is 30;
[0028] b. The depth of SNP site sequencing, that is, the DP value in the vcf file, the default is 10;
[0029] c. Whether to consider the case of missing sequencing, that is, the case where the genotype in the vcf file is . / ., it is not considered by default.
[0030] (3) Define specific criteria;
[0031] That is, set the values of thresholds A and B...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com