Yarrowia lipolytica for the production of farnesene
A technology for producing farnesene and bacteria, applied in the field of genetic engineering, can solve the problems of enhancing precursor flow, weakening branched metabolic pathways, and low fermentation level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
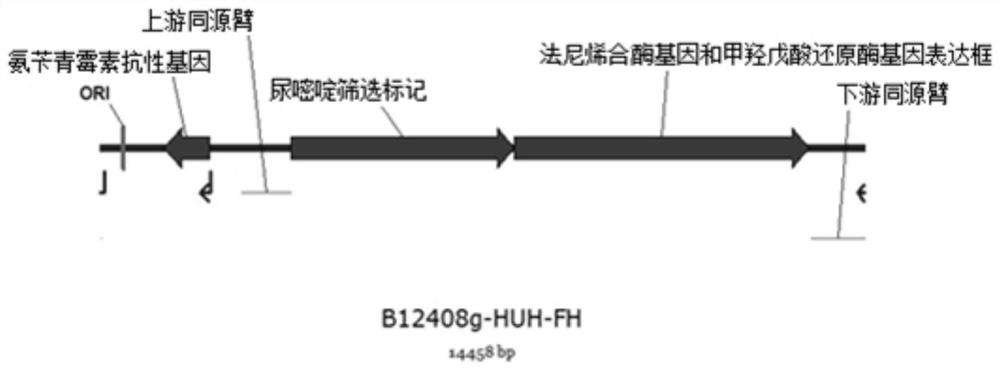
[0041] Example 1: Construction of Backbone Plasmid B12408g-HUH
[0042] The primer sequences used in the construction of the backbone plasmid B12408g-HUH include:
[0043] 12408g-F: 5'-ATTTAAAT GATTGAGACGTGGCGATCCGGCAAC-3' (SEQ ID NO: 2),
[0044] 12408g-R: 5'-ATTTAAAT TAAGAAGCACACCACAGAACCGTAC-3' (SEQ ID NO: 3);
[0045] 408g-F:
[0046] 5'-CCCAAAGAGGTGAATTCGGCAAGCTTCTAGAGTAGGTGGTATGTACTCGTACTGTACT-3' (SEQ ID NO: 4),
[0047] 408g-R:
[0048] 5'-CTACTCTAGAAGCTTGCCGAATTCACCTCTTTGGGCCATTTCATTAGATCG-3' (SEQ ID NO: 5).
[0049] "-F" in the above names stands for forward; "-R" stands for reverse.
[0050] Using the CIBTS2112 genome as a template, the upstream and downstream homology arms were amplified by PCR using primer sets 12408g-F / 408g-R and 408g-F / 12408g-R, respectively. PCR conditions: Denaturation at 95°C for 5min, denaturation at 95°C for 30s, annealing at 58°C for 30s, extension at 68°C for 2min, 32 cycles, 68°C for 10min, and finally 16°C for 10min.
[0051] Usin...
Embodiment 2
[0053] Example 2: Construction of plasmid B12408g-HUH-FH
[0054] 1.1 Preparation of pTEF-FS-xpr2t expression module
[0055] Using the plasmid pUC57-FS as a template, the pTEF-FS-xpr2t expression module was obtained by PCR amplification using the following 408-FH-F / T-FS-X-R primer pair.
[0056] 408-FH-F:
[0057] 5'-TACCCTGATTGACTGGAACAGCCCCAAGAGACCGGGTTGGCGGCGTATTTGTG-3' (SEQ ID NO: 6),
[0058] T-FS-X-R:
[0059] 5'-GGTGCACCGAAGAGCCTGTGGAAGCGACGGGGACACGGGCATCTCACTTGC-3' (SEQ ID NO: 7).
[0060] PCR conditions: Denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 58°C for 30 seconds, extension at 68°C for 3 minutes, 32 cycles, incubation at 68°C for 10 minutes, and finally incubation at 16°C for 10 minutes.
[0061] 1.2 Preparation of pEXP-tHmgR-pex20t expression module
[0062] The plasmid TAs-7h-GH was used as a template to obtain the pEXP-tHmgR-pex20t expression module by PCR amplification using the following E-H-P-F / 408-FH-R primer ...
Embodiment 3
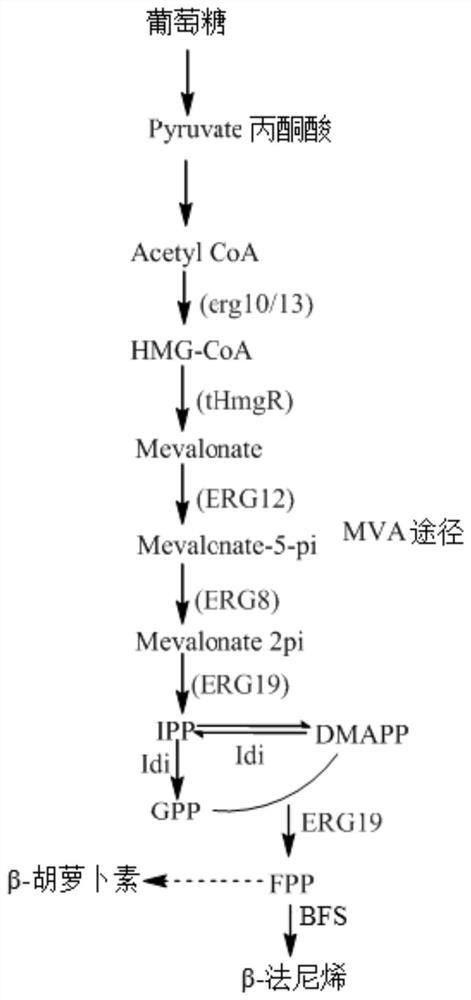
[0069] Example 3: Construction of β-farnesene-producing bacteria
[0070] The plasmid B12408g-HUH-FH constructed in Example 2 was digested with SwaI to recover a large fragment, and the ZymogenFrozen-EZ Yeast Yransformation kit II kit (Zymo Research Corporation) was used to transform CIBTS2112 to obtain the genetically engineered strain CIBTS2617.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com