Traceless deletion strain DPV CHv-gEdeltaCT in ET region of gE gene of duck plague virus and construction method of traceless deletion strain
A technology of duck plague virus and construction method, which is applied in the field of genetic engineering and can solve problems such as residual bases and residual MiniF elements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
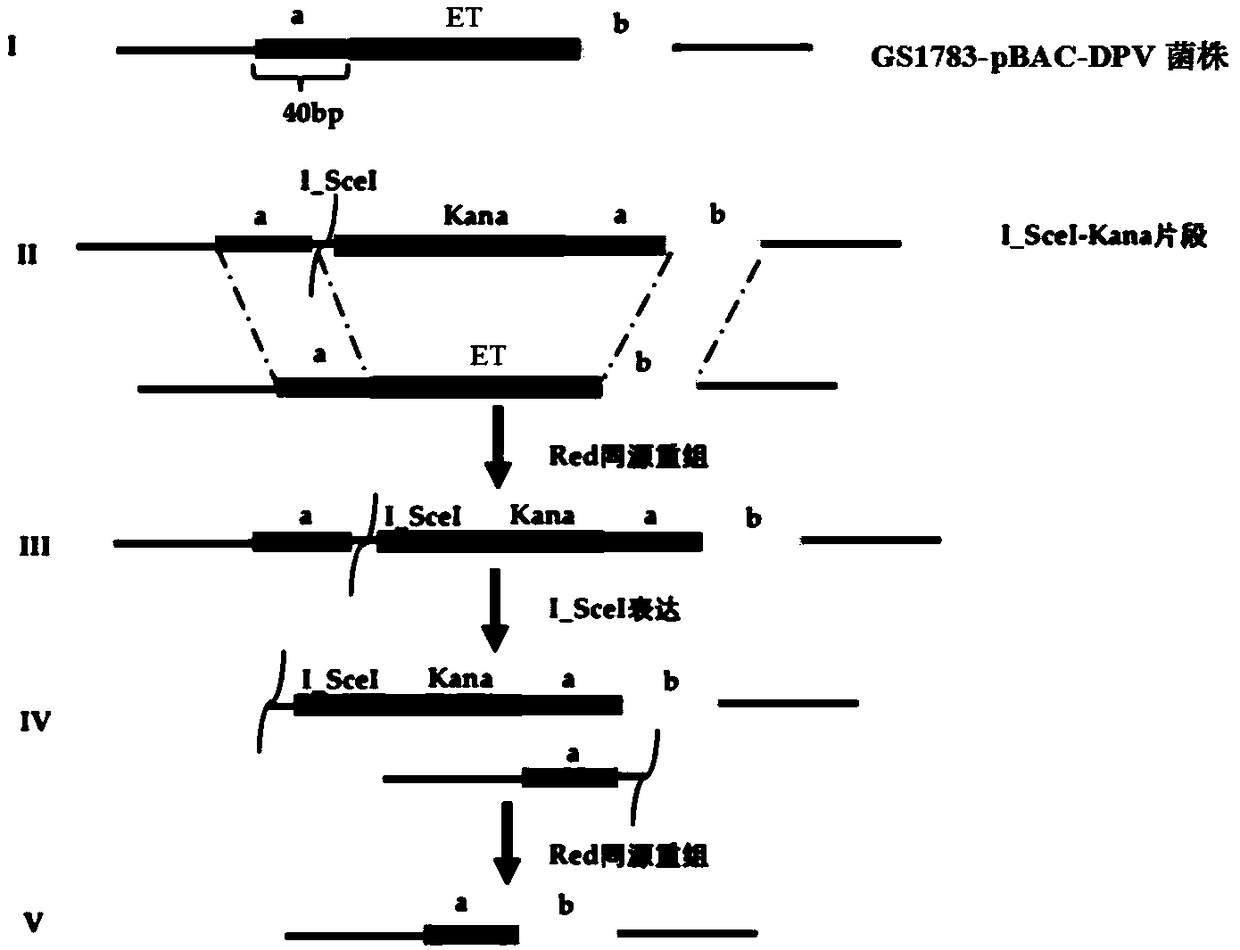
[0051] Example 1 Preparation of duck plague virus gE gene ET region seamless deletion strain DPV CHv-gEΔET
[0052] Duck plague virus gE gene ET region traceless deletion strain DPV CHv-gEΔET, its construction method comprises the following steps:
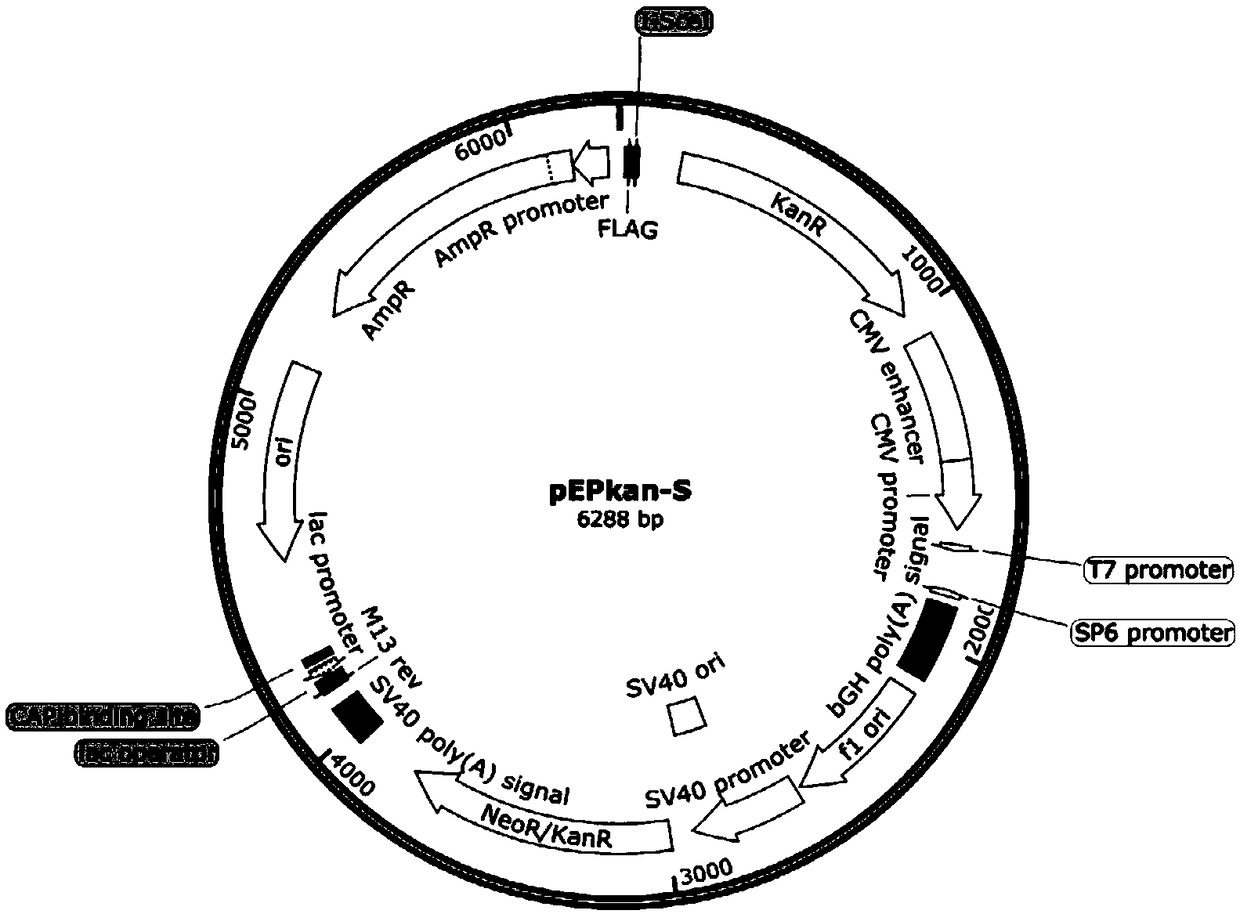
[0053] 1. Prepare GS1783 electroporation competent, electrotransform pBAC-DPV plasmid
[0054] (1) Escherichia coli with pBAC-DPV plasmid was revived in LB solid medium containing chloramphenicol, cultured overnight at 37°C; single colonies were inoculated in LB liquid medium containing chloramphenicol, and cultivated overnight at 37°C;
[0055] (2) Extract the pBAC-DPV plasmid according to the operating instructions of the QIAGEN Plasmid Midi Kit;
[0056] (3) Resuscitate GS1783 frozen-preserved bacteria in LB solid medium, culture overnight at 30°C;
[0057] (4) Pick a single colony of GS1783 and inoculate it in 5 mL of LB liquid medium, and culture it overnight at 30°C to obtain a seed solution;
[0058] (5) Add 5mL seed liqu...
Embodiment 2
[0160] Example 2 Determination of the growth curve of DPV CHv-gEΔET of the traceless deletion strain
[0161] 1. Determination of one-step growth curve
[0162] The parental virus DPV CHv and gE gene ET region traceless deletion strain DPV CHv-gEΔET were inoculated into DEF cells at 2 MOI, and the supernatant and cells were collected at 6h, 12h, 18h, and 24h after inoculation, and each time point was repeated three times. After the collection was complete, the freeze-thaw was repeated twice, and the virus titer was detected in a 96-well plate. The results showed that the deletion of the ET region of the gE gene affected the replication of the DPV CHv virus.
[0163] 2. Determination of multi-step growth curve
[0164] The parental virus DPV CHv and gE gene ET region traceless deletion strain DPV CHv-gEΔET were inoculated into DEF cells at a MOI of 0.01, and the supernatant and cells were collected 12h, 24h, 48h, and 72h after inoculation, and each time point was repeated thre...
Embodiment 3
[0165] Example 3 Plaque formation experiment of DPV CHv-gEΔET strain without trace deletion
[0166] Inoculate the parental virus DPV CHv and gE gene ET region traceless deletion strain DPV CHv-gEΔET at 0.01 MOI to a 6-well plate filled with DEF cells, 37°C, 5% CO 2 After absorbing for 2 hours, remove the supernatant, add 2 mL of 1% methylcellulose, 37 ° C, 5% CO 2 After culturing for 48 h, remove 1% methylcellulose, wash 3 times with PBS, fix overnight at 4°C with 4% paraformaldehyde, wash 3 times with PBS, add H 2 o 2 The mixture mixed with methanol at a volume ratio of 1:50 was incubated at room temperature for 30 minutes, washed 3 times with distilled water, added 5% BSA blocking solution, incubated at room temperature for 30 minutes, added rabbit anti-DPV, incubated overnight at 4°C, washed 3 times with PBS, added Biotinylated goat anti-rabbit IgG, incubated at 37°C for 30 minutes, washed 3 times with PBS, added dropwise SABC substrate, incubated at 37°C for 30 minutes,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com