Accurate single-molecule force spectroscopy method with high throughput
A single-molecule force spectroscopy, high-throughput technology, applied in the field of precise high-throughput single-molecule force spectroscopy, can solve the problems of low detection efficiency, poor specificity and accuracy, and achieve high detection efficiency, specificity and accuracy Enhanced, efficient and precise measurement of the effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
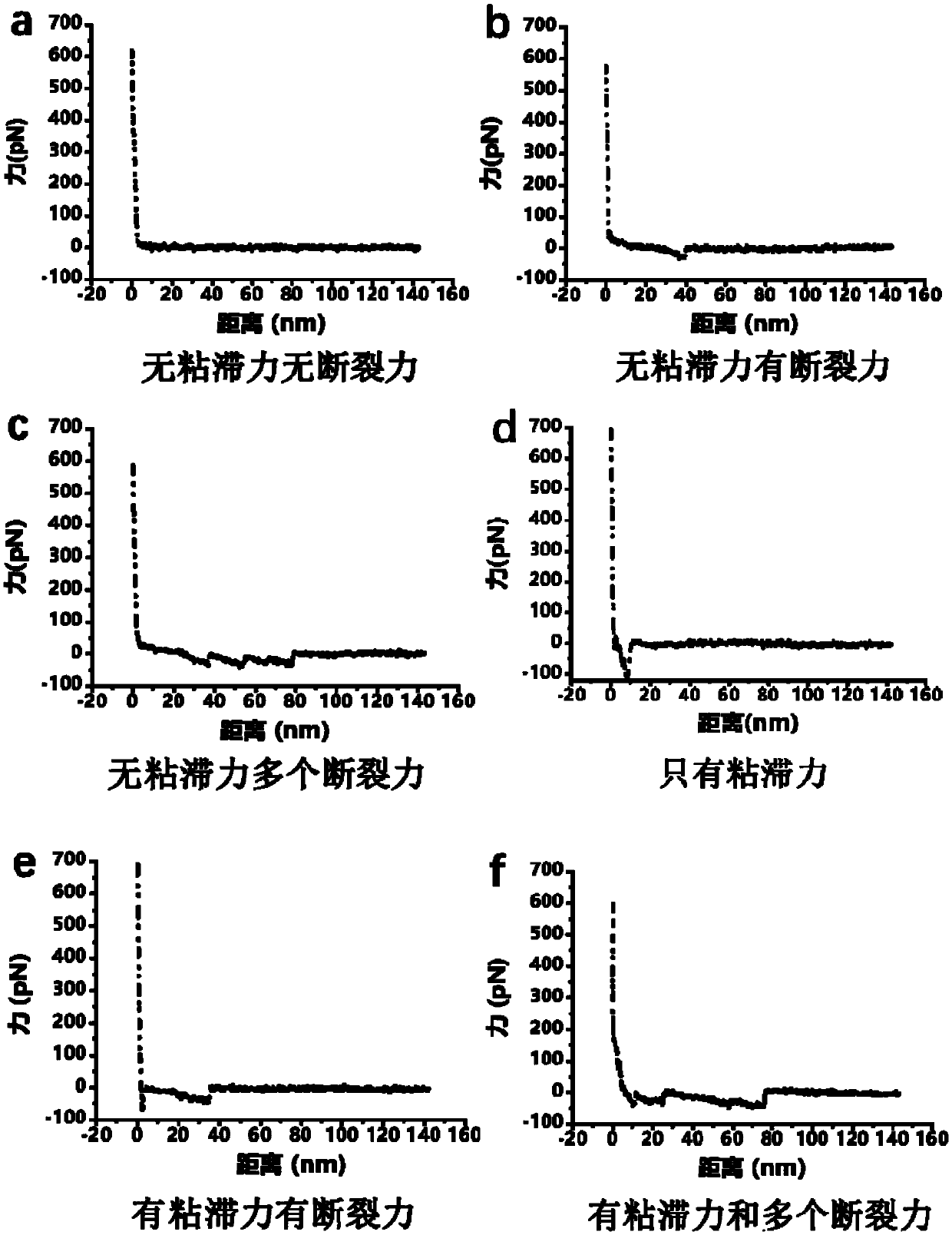
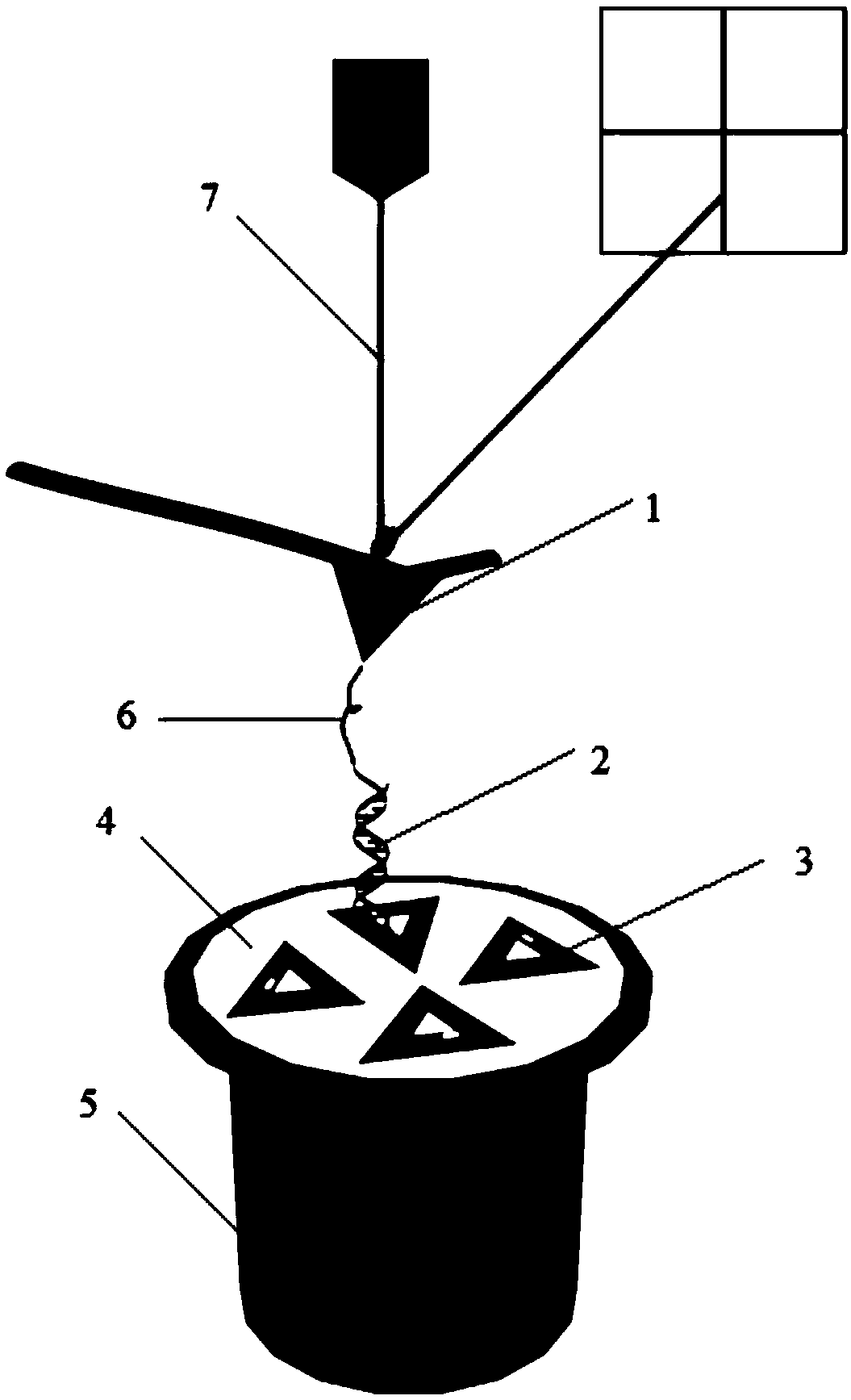
[0039] The DNA origami used in this example is a triangular DNA origami, which can be self-assembled into a triangular pattern with an external side length of 120 nm and an internal side length of 40 nm by using M13mp18 DNA and staple strands and a single strand of target DNA staple. That is, a DNA origami modified with single-strand DNA staples of interest. On the three sides A, B, and C of the triangle DNA origami, six modification sites were respectively selected, specifically as figure 2 As shown, a target DNA short chain is extended at the 5' end of the oligonucleotide chain at the selected modification site. The target DNA short chain can be 20 bases, which can be as short as the DNA immobilized on the AFM needle tip. The strands are complementary and paired, and then the interaction force between the two short DNA strands is measured by single-molecule force spectroscopy using an atomic force microscope.
[0040] 1.1 Preparation of triangular DNA origami with target D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com