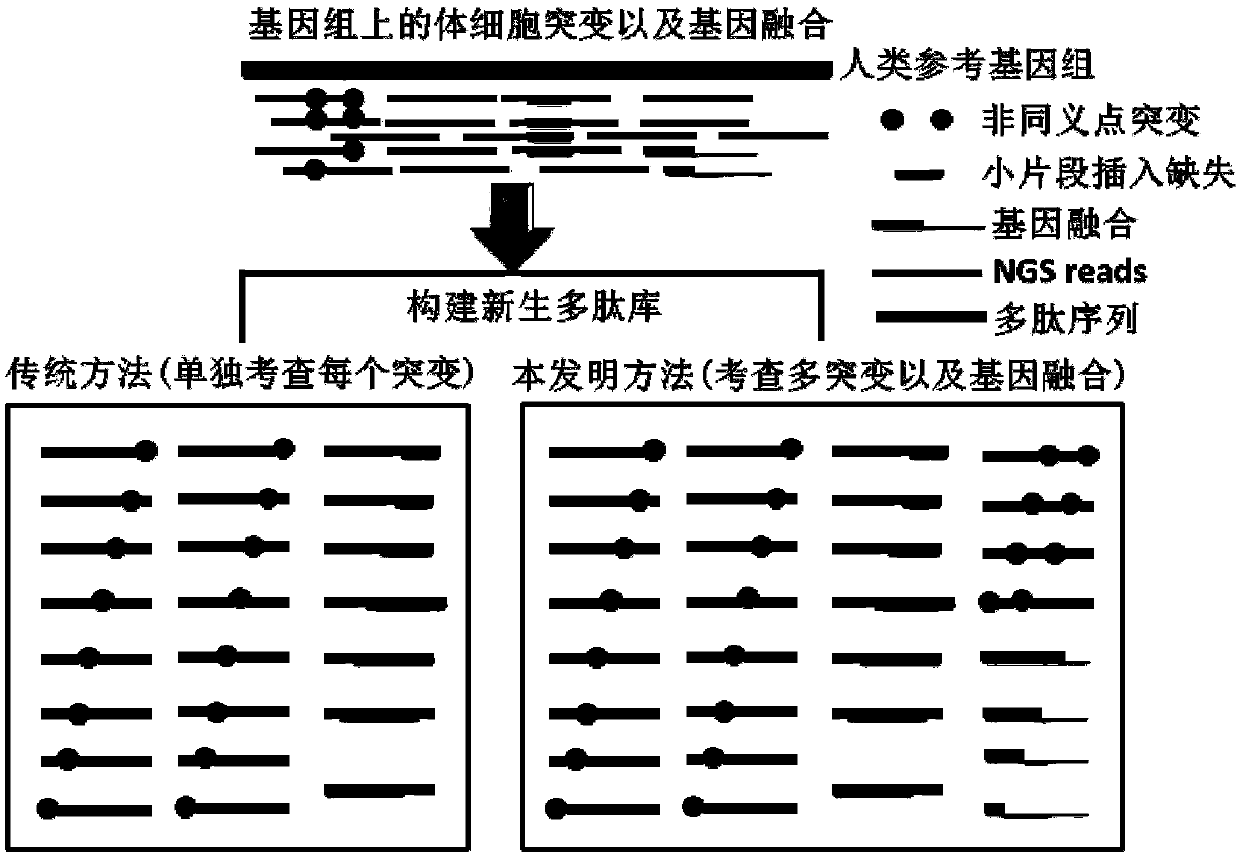
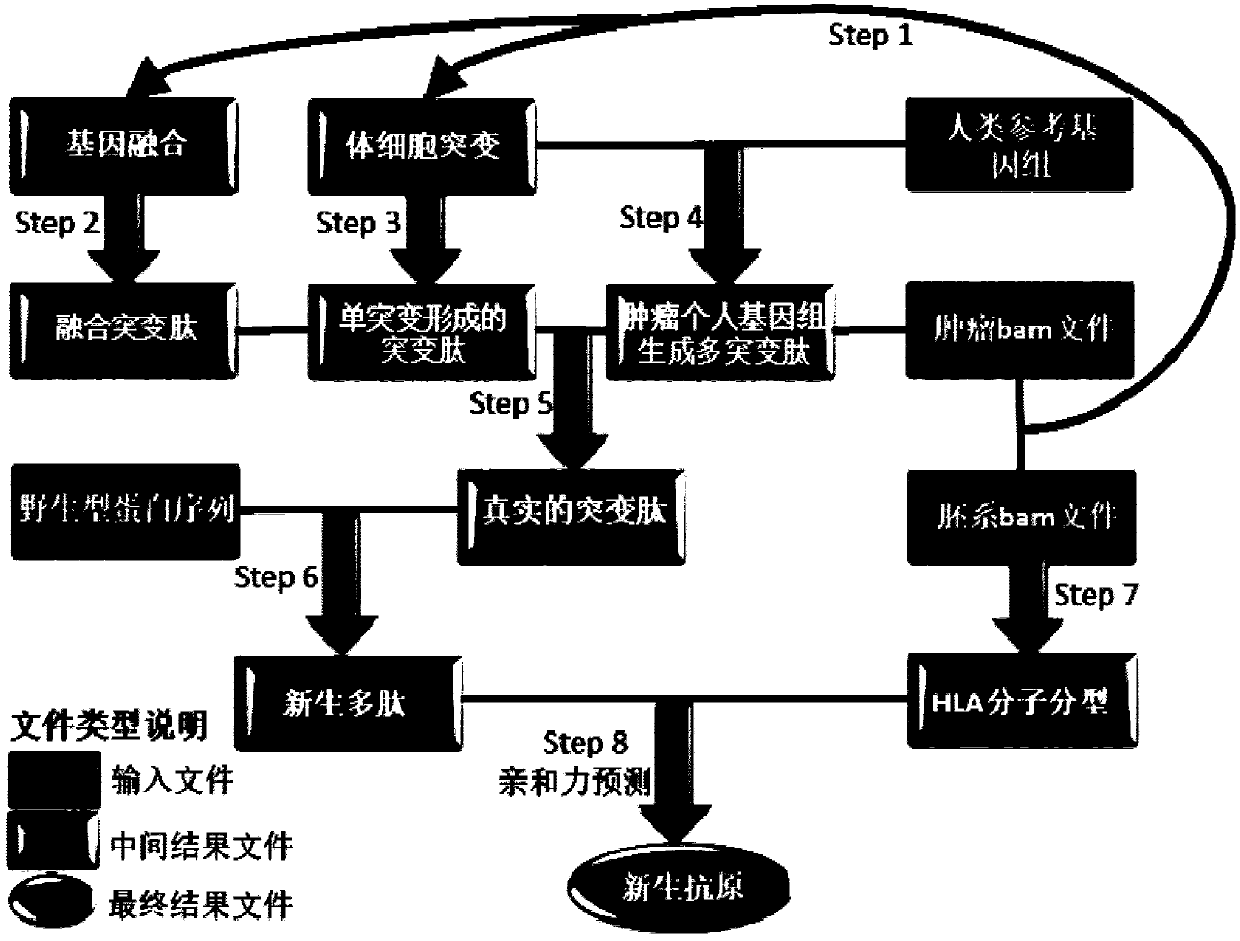
Method and device for predicting tumor newly-born antigen and storage medium
A neonatal and tumor technology, applied in the field of bioinformatics, can solve problems affecting the accuracy of neoantigen prediction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
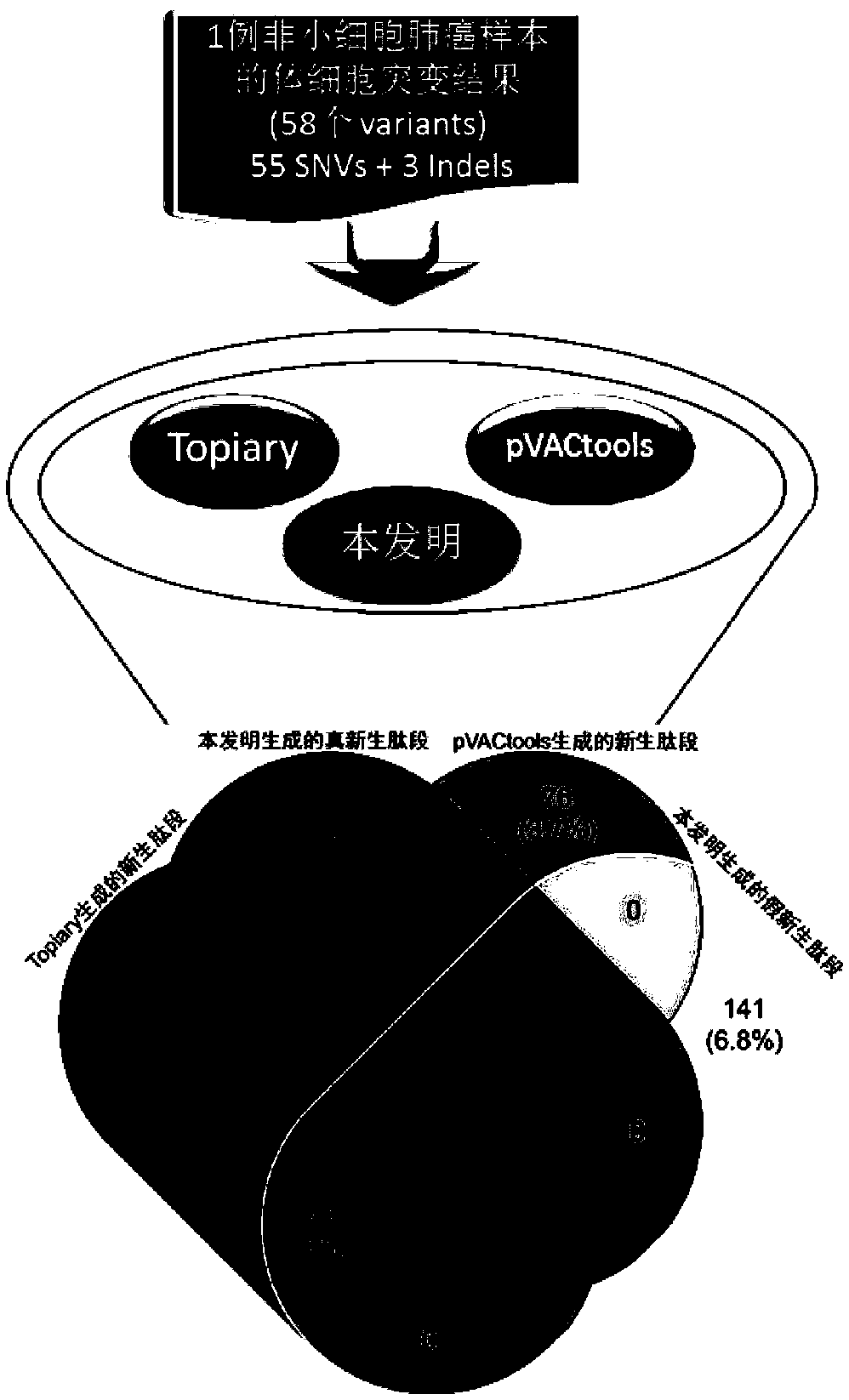
[0093] This embodiment starts with the mutation file of a sample of non-small cell lung cancer. The specific mutation information is shown in Table 1, and Topiary, pVACtools and the method of the present invention are compared to predict the nascent polypeptide with a length of 8-11 amino acids.
[0094] This embodiment can be an implementation example of the verification scheme of the present invention, proving the advantages of the present invention compared with the two current mainstream tools. image 3 The comparison process and results between the neoantigen prediction method of the present invention and two mainstream open source tools are illustrated. Since the three tools use the same method for the affinity of peptides and HLA molecules, we only focus on the differences between the three tools in the generation of nascent peptides.
[0095] Table 1 Information on 58 somatic mutation sites in patients with non-small cell lung cancer
[0096]
[0097]
[0098] I...
Embodiment 2
[0104] Example 2 is a specific application scenario of the method provided by the present invention in tumor immunotherapy, to illustrate the application value of the present invention in tumor immunotherapy, and its advantages compared with the currently approved tumor mutation load. High tumor mutational burden indicates that there are more tumor somatic mutations, which means that more tumor neoantigens can be produced, so that the tumor cells are more likely to be recognized by immune cells, which is why tumor mutational burden is used as a biomarker The biological rationale for assessing the effects of immunotherapy. Example 2 verifies the effectiveness of the tumor neoantigen load calculated by the present invention as a biomarker for immunotherapy.
[0105] Table 3 lists the overall survival data of 13 cases of hepatocellular carcinoma patients undergoing immunotherapy, as well as the tumor mutation load detected by whole exome sequencing, and the sample neoantigen load...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com