Method for detecting blood microsatellite instability based on second generation sequencing technology
A technology of microsatellite instability and next-generation sequencing technology, applied in sequence analysis, instruments, hybridization, etc., can solve the problem of blood samples not being very good, and achieve high consistency results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
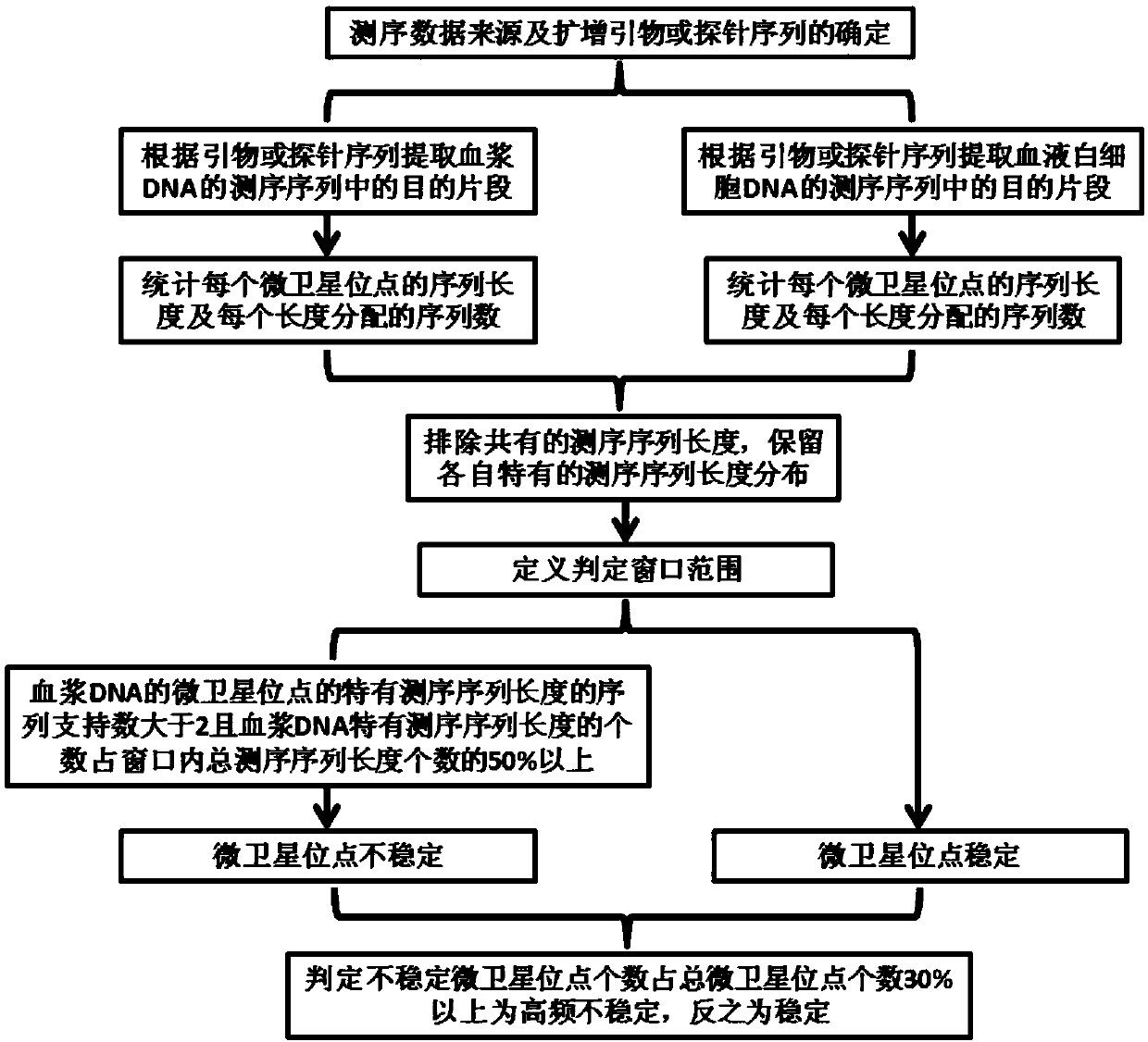
[0050] Select 10 cases of corresponding plasma and white blood cell samples known to have passed the 3730 Yuewei kit (patent number: ZL 2011 1 0152226.X) to verify the results of MSI (3 cases of MSI-H high-frequency unstable type, 7 cases of MSS stable type ) of the next-generation amplicon sequencing data, the following detection methods were performed.
[0051] The specific detection method is illustrated by taking an example sample1 of a high-frequency unstable MSI-H as an example.
[0052] 1. Determine the 5 biomarkers of the microsatellite loci used in the data, namely BAT25, BAT26, MONO27, NR21 and NR24, and determine the corresponding amplification primer sequences for the 5 markers, as follows:
[0053] BAT25 primers:
[0054] Primer 1: TCTGCATTTTAACTATGGCTC (SEQ ID NO: 1)
[0055] Primer 2: CTCGCCTCCAAGAATGTAAGT (SEQ ID NO: 2)
[0056] BAT26 primer:
[0057] Primer 1: CTGCGGTAATCAAGTTTTTAG (SEQ ID NO: 3)
[0058] Primer 2: AACCATTCAACATTTTTTAACCC (SEQ ID NO: 4) ...
Embodiment 2-10
[0079] Examples 2-10 were selected from the corresponding plasma and white blood cell samples (3 cases of MSI-H high-frequency non Stable type, the second-generation amplicon sequencing data of 7 cases of MSS stable type) were used as objects, and the above steps 1-10 were repeated. The results are shown in Table 1.
[0080] Table 1
[0081] Sample ID
[0082] Note: MSI-PCR verification results and prediction results statistics, where "+" means unstable, "-" means stable.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com