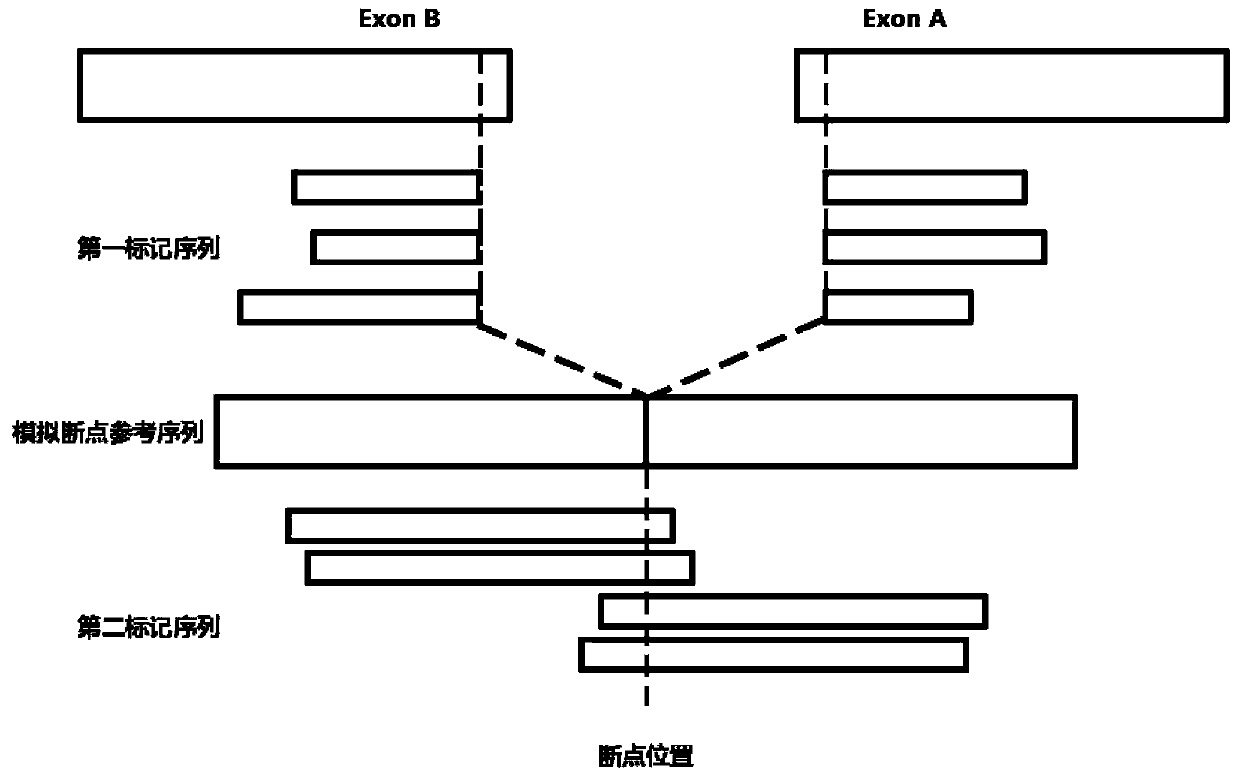
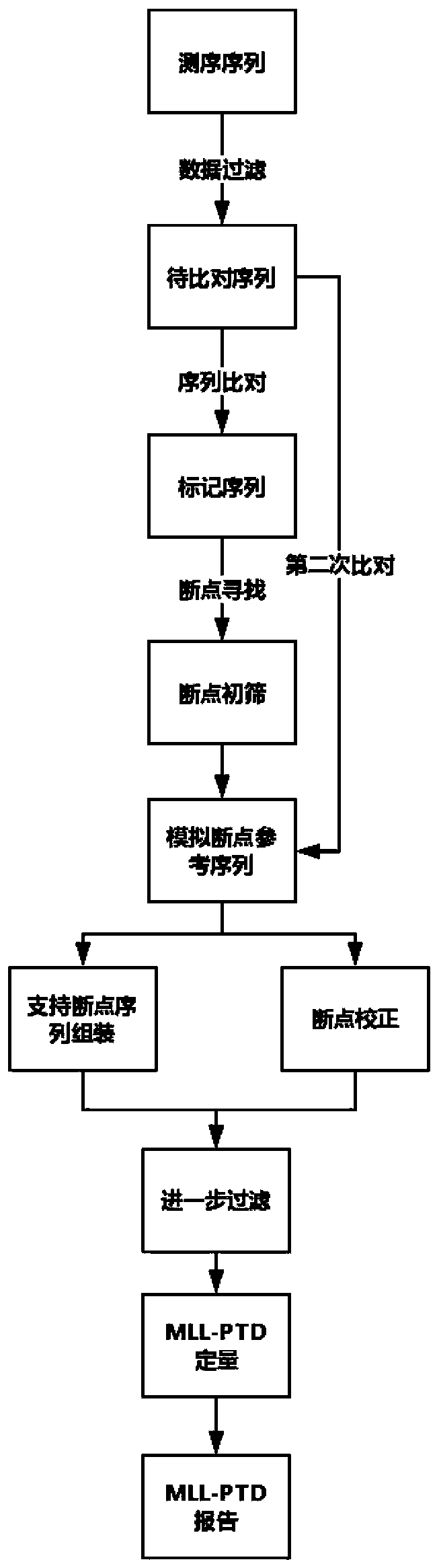
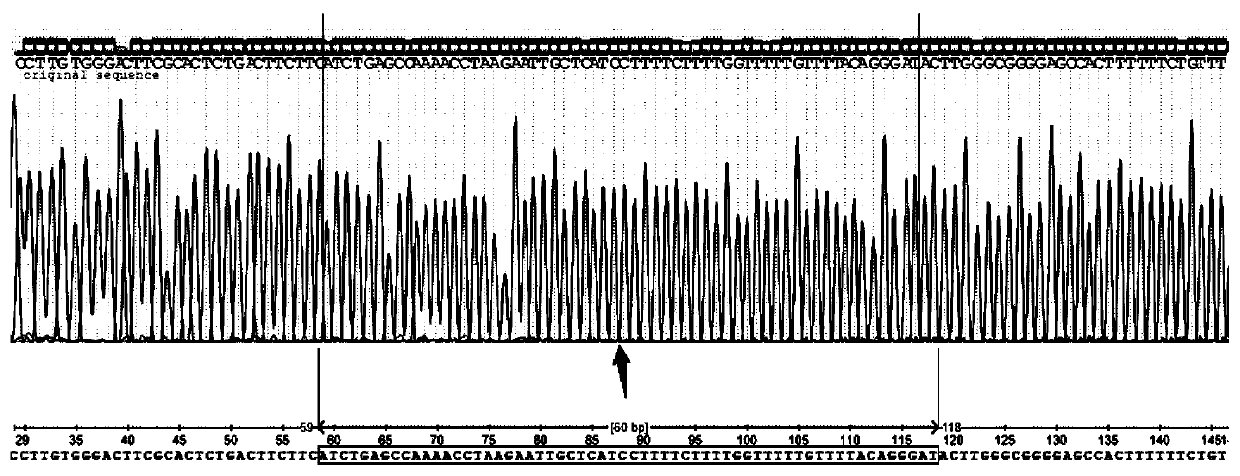
Method for detecting gene rearrangement, device, storage medium and processor
A technology of gene rearrangement and storage medium, applied in the fields of genomics, instruments, proteomics, etc., can solve the problem of difficulty in detecting the position of gene rearrangement breakpoints, and achieve the effect of low cost and high detection accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1 detects the method for MLL-PTD gene rearrangement
[0063] 1. Samples and data
[0064] 1) Extract the patient's bone marrow or peripheral blood and store it in a collection tube.
[0065] 2) Extract the sample nucleic acid, and store the remaining samples at -80°C.
[0066] 3) Construct a sequencing library, and enrich the target region by hybridization capture method (the hybridization capture probes of the MLL gene are 36 exons of the gene, see Table 1 below for the specific sequence).
[0067] 4) The captured library is sequenced on the machine.
[0068] Table 1:
[0069]
[0070]
[0071]
[0072]
[0073]
[0074]
[0075] 2. Preprocessing of sequencing data
[0076] 1) Data quality control
[0077] Mainly to delete low-quality sequences, sequences containing more than 5 bases N were deleted; sequences with an average sequencing quality of 40 consecutive nucleotides lower than Q20 were also deleted.
[0078] 2) Alignment of MLL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com