MiRNA-disease association predicting method based on double random walk models
A prediction method, a double-random technology, applied in the interdisciplinary field of bioinformatics and artificial intelligence, can solve problems such as time-consuming, prediction accuracy needs to be improved, waste of manpower and financial resources, etc., and achieve the effect of rich information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
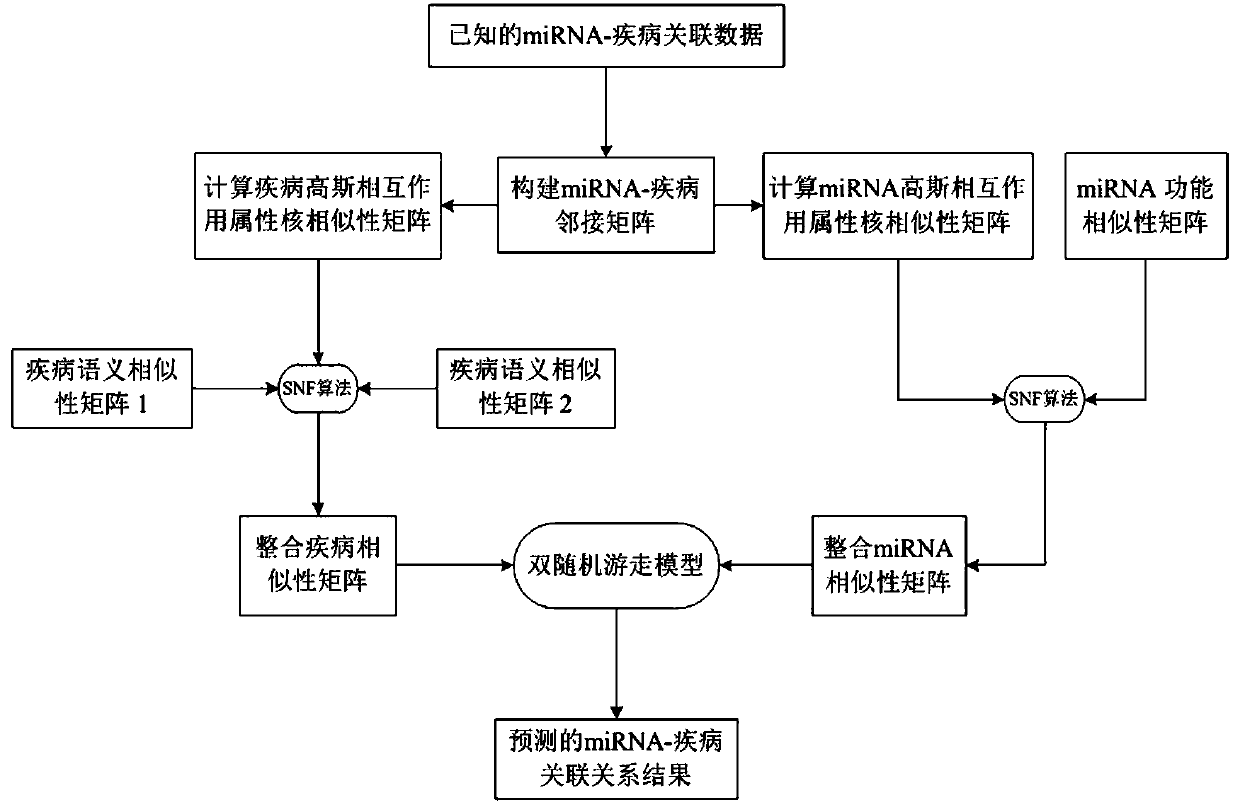
[0055] refer to figure 1 , a miRNA-disease association prediction method based on a double random walk model, comprising the following steps:
[0056] 1) Obtain the miRNA-disease association data set and construct an adjacency matrix about the miRNA-disease association: obtain the miRNA-disease association data confirmed by biological experiments from the HMDD database, and obtain 5430 pairs of different miRNA and disease association data, which involve disease There are 383 kinds of miRNAs and 495 kinds of miRNAs. Define D={d(1), d(2), d(3),...,d(nd)} to record the set of nd diseases, M={m( 1), m(2), m(3),..., m(nm)} to record the set of nm miRNAs, and build an adjacency matrix MD na×nm Represents the relationship between miRNA and disease association data, when disease d(i) and miRNAm(j) are verified as association, the adjacency matrix MD nd×nm The value of MD(i, j) is set to 1; otherwise, the value of MD(i, j) is set to 0, indicating an unknown association;
[0057] 2)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com