Replication system and application thereof in gene expression
A gene and replicase technology, applied in bacteria, fermentation, etc., can solve the problems of RNA replication speed and replicon stability, unfavorable long-term retention of RNA replicon, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] Example 1 Verification of replication efficiency when replicase and replication sequences are expressed separately
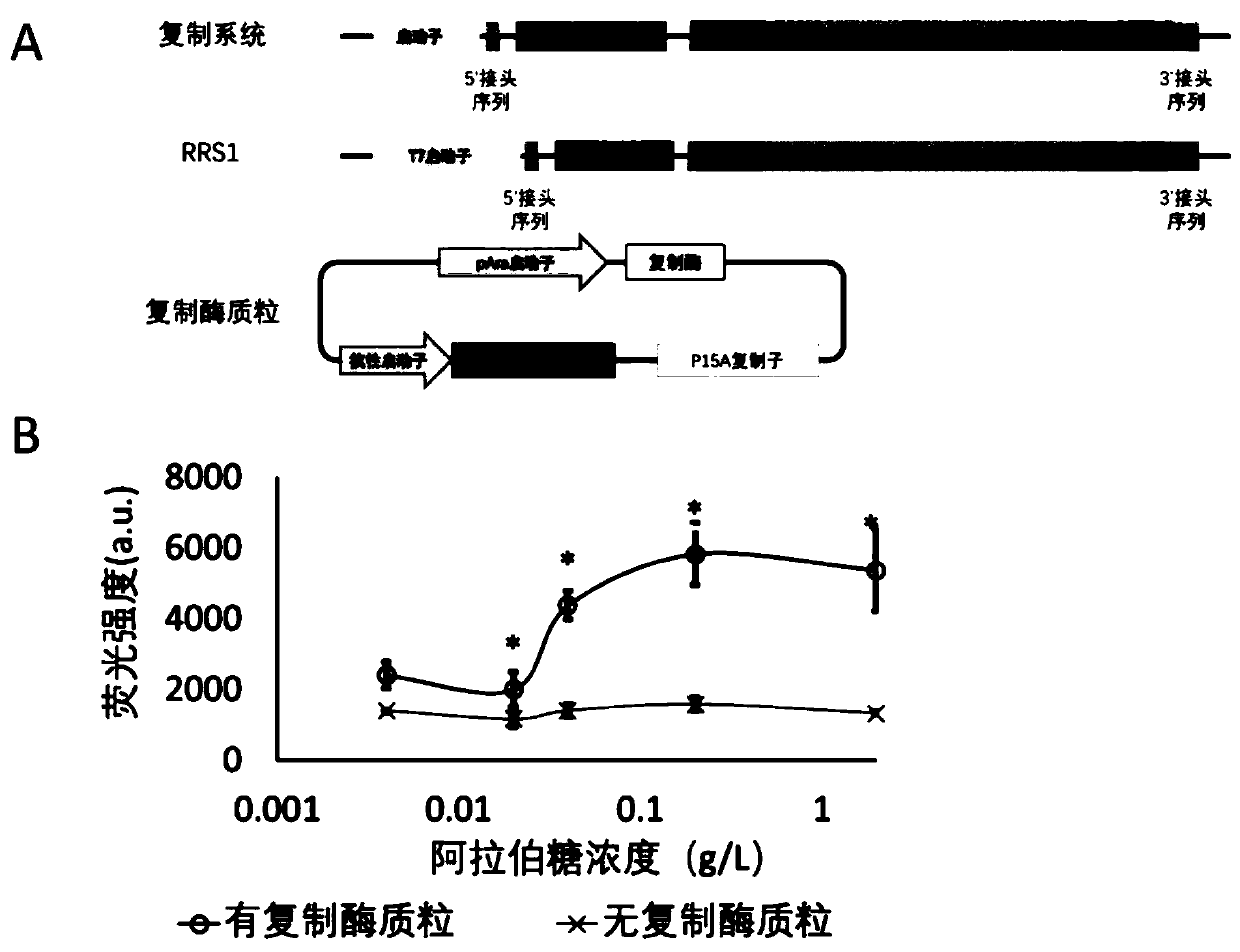
[0103] The design of the replication system for expressing the replicase and replicating sequences separately is as follows: figure 1 As shown in the schematic diagram of A, the replication sequence and replicase are expressed separately, the replication system expresses the replication sequence, but not the replicase, and the replicase is expressed in another replicase plasmid. The replication sequence contained in the replication system includes the promoter sequence, the 5' linker sequence, the target gene sequence (that is, the "target protein region" in the figure), and the inactive replicase sequence (that is, the target protein region) from the 5' end to the 3' end. "phage sequence (replicase inactive)") and 3' linker sequence, such as figure 1 Shown in "Replication System" in A. Since the replicase in the replication system is inactivated, the...
Embodiment 2
[0107] Example 2 Construction of preferred RNA replication system and detection of its working effect
[0108] (1) construct the plasmid of preferred RNA replication system
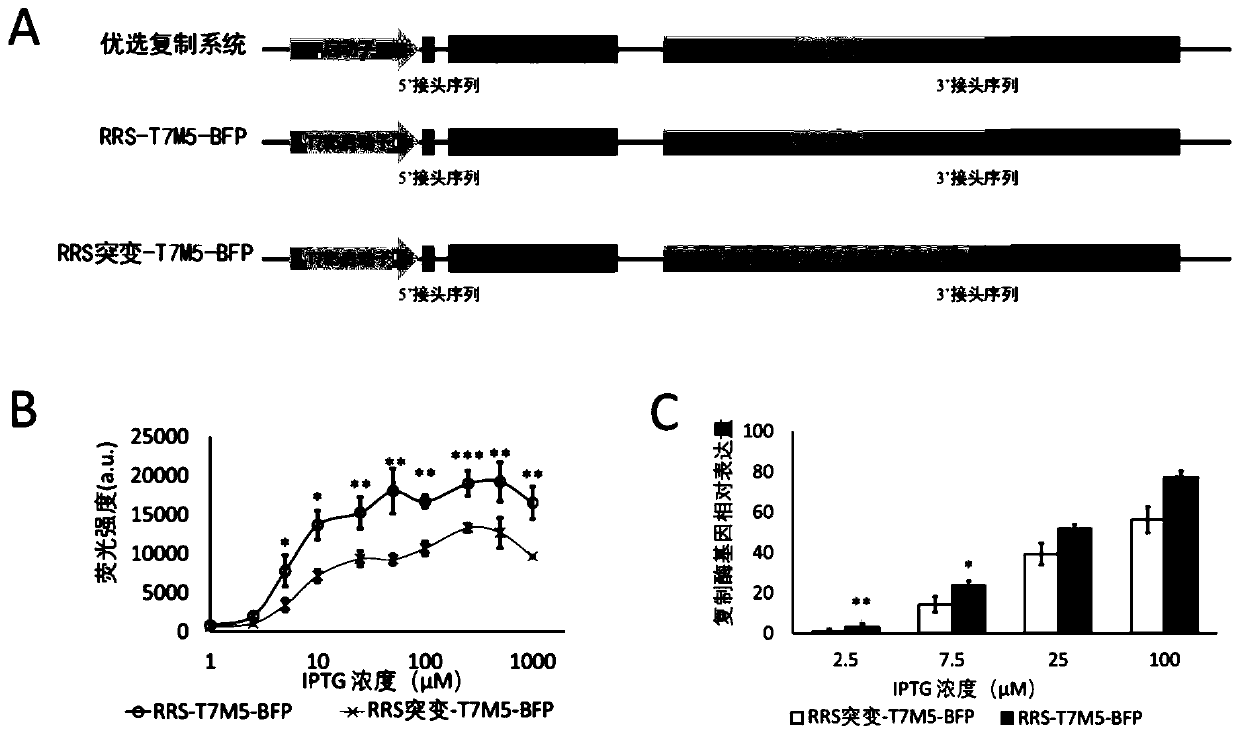
[0109] A schematic diagram of the preferred replication system is shown in figure 2 As shown in "preferred replication system" in A, the replicase sequence is placed in the replication system and co-expressed with the target gene. The preferred RNA molecule of the replication system comprises a promoter, a 5' linker sequence, an objective gene sequence (i.e. the "target protein region" in the figure), a replicase sequence (i.e. the "Qβ replicase" in the figure), a 3' linker sequence and HDV ribozyme sequence, wherein the HDV ribozyme sequence is used for self-cleavage after expression.
[0110] In this embodiment, the blue fluorescent protein BFP is used as the target gene, and the T7M5 promoter is used to transcribe the RNA molecule of the replication system. The schematic diagram of the replication...
Embodiment 3
[0133] Embodiment 3 cuts the upstream and downstream of the replication system
[0134] In this embodiment, HDV ribozyme and tRNA were used to cut the 3' end and 5' end of the replication system, respectively, to test the impact of removing the remaining sequences outside the 3' end and 5' end on the performance of the replication system.
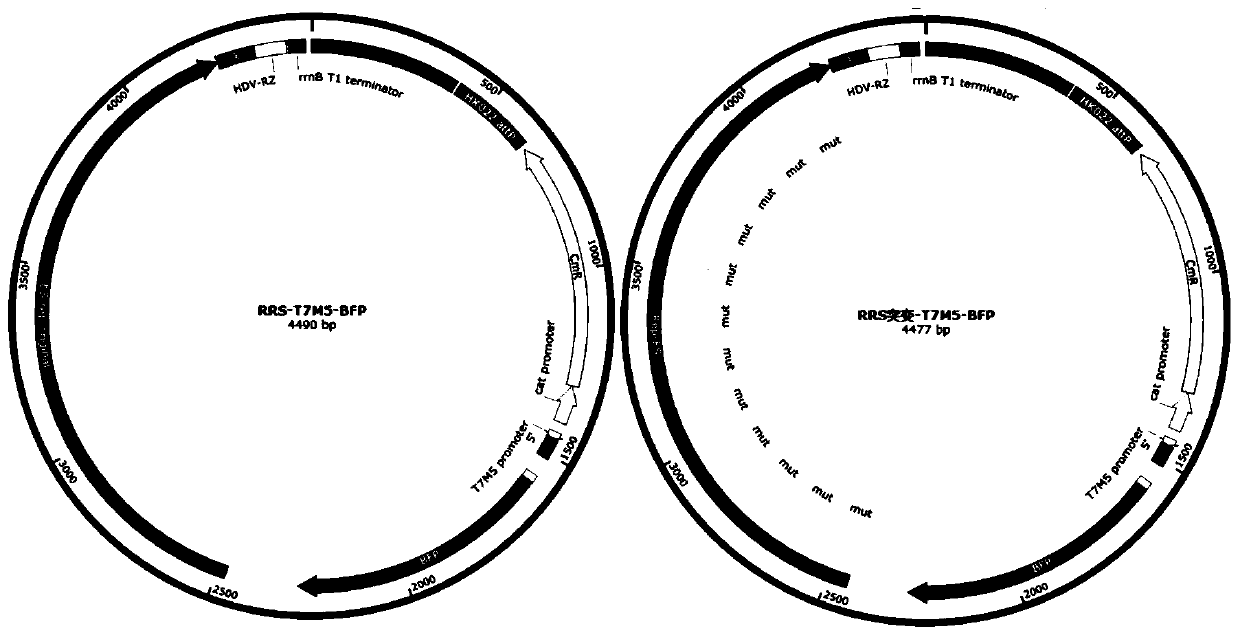
[0135] (1) Use HDV ribozyme to cut the 3' end of the replication system, the schematic diagram is as follows Figure 4 As shown in A, the replication system RRS-T7M5-BFP constructed in Example 2 and the control replication system RRS mutation-T7M5-BFP were used. In addition, the replication system RRS-T7M5-BFP and the control replication system RRS mutation-T7M5-BFP not containing the HDV ribozyme sequence after the 3' linker sequence were constructed.
[0136] Using the same plasmid construction method as in Example 2 and the attB / P integration method of HK022, the plasmids expressing these replication systems were integrated into the g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com