Prediction method of miRNA target genes based on convolutional neural network
A target gene prediction, convolutional neural network technology, applied in the field of bioinformatics, can solve problems such as data set imbalance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
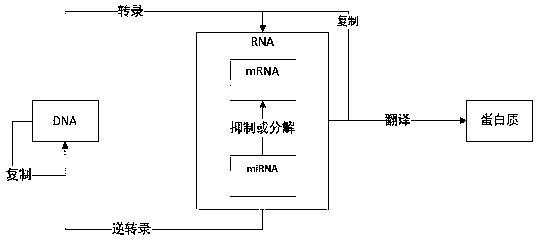
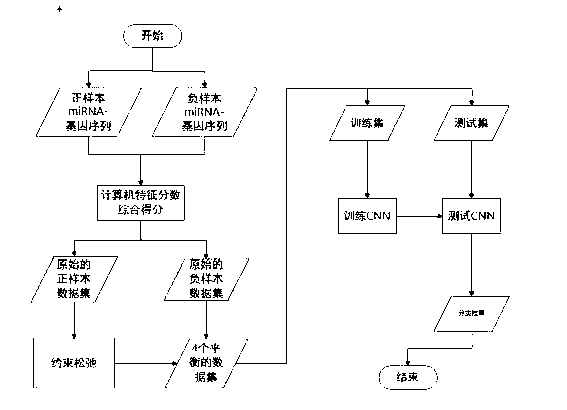
[0036] Such as Figure 1-4 Shown, a kind of miRNA target gene prediction method based on convolutional neural network, comprises the following steps:
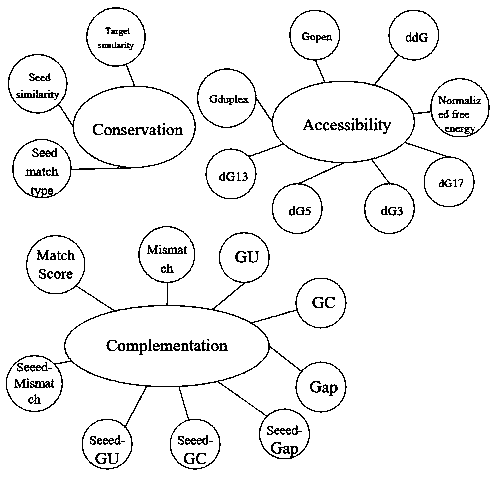
[0037] S1: According to the published miRNA-mRNA pair, download the corresponding sample data mRNA required for the experiment from the NCBI library, download the corresponding sample data miRNA required for the experiment from the miRBase library, and calculate the eigenvalues of the original and negative samples, where , the characteristics are divided into three categories: complementarity, accessibility, and conservation; and complementarity can be evaluated from 9 characteristics, 9 characteristic values; accessibility can be evaluated from 8 characteristics, 8 Eigenvalues; Conservatism is evaluated from 3 eigenvalues, 3 eigenvalues; therefore a total of 20 eigenvalues need to be calculated;
[0038] S2: Construct a balanced data set: In order to obtain more candidate sites, set loose thresholds for the three characte...
Embodiment 2
[0053] Concrete steps of the miRNA target gene prediction method based on convolutional neural network of the present invention:
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com