Reverse transcription PCR method mediated by terminal deoxynucleotidyl transferase and used for detecting micro RNA
A deoxynucleotide and transferase technology, applied in biochemical equipment and methods, microbial assay/inspection, etc., can solve the problems of inability to catalyze the polymerization of deoxyribose triphosphate, short targets, etc., and achieve good detection specificity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
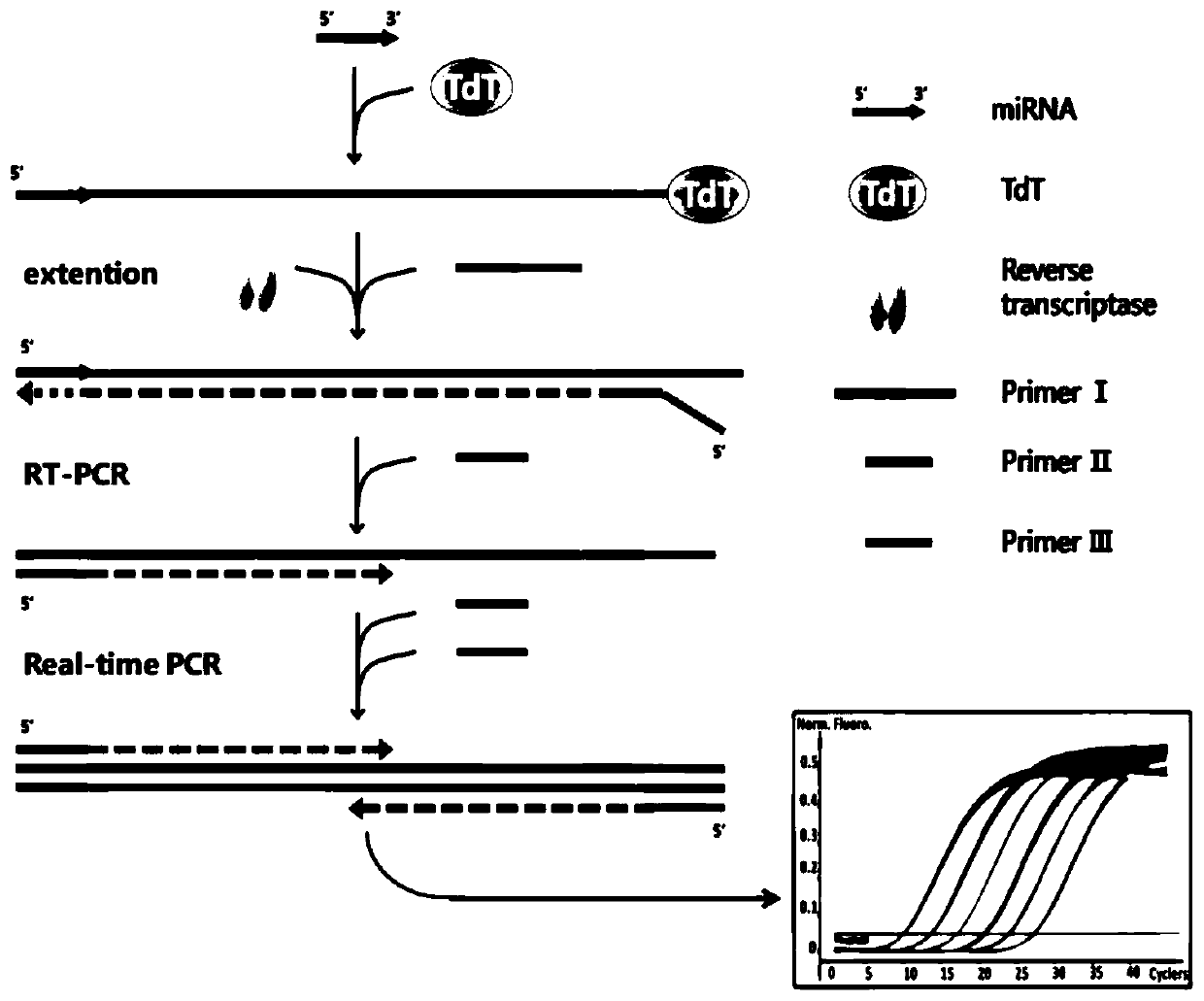
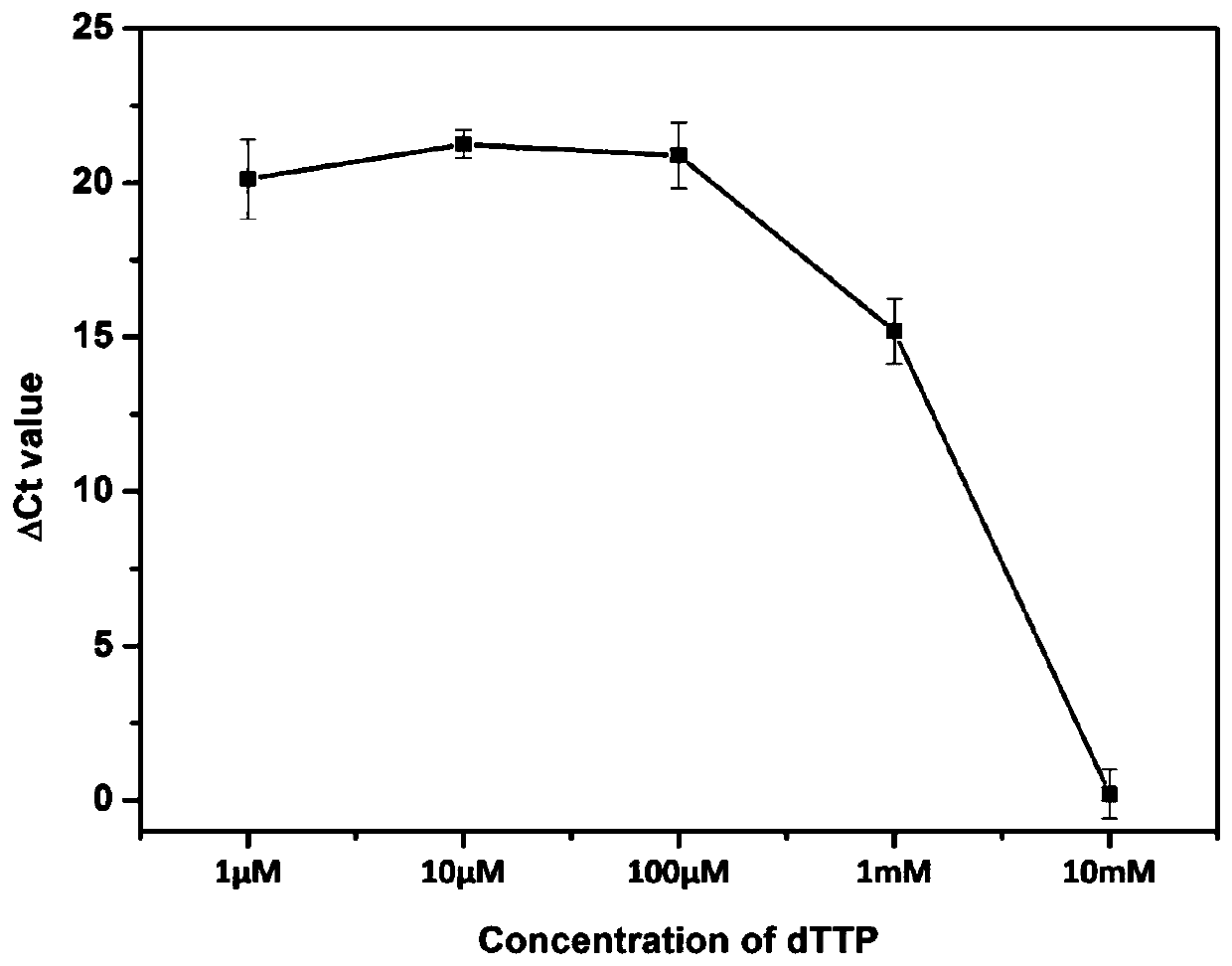
[0060] (1) TdT extension step: first prepare 2 , 10pM DNA-122 (sequence shown in SEQID No.4), 1×TdT reaction buffer and 10U TdT enzyme reaction system, then react at a constant temperature of 37°C for 30min, and then inactivate at 85°C for 25min.
[0061] (2) Reverse transcription step: Prepare a reaction system containing 10-fold diluted reaction product from the previous step, 100 nM primer I (sequence shown in SEQ ID No.1), 1× reverse transcription buffer, and reverse transcriptase MixI, Then react at a constant temperature of 37°C for 15min, followed by inactivation at 85°C for 5s.
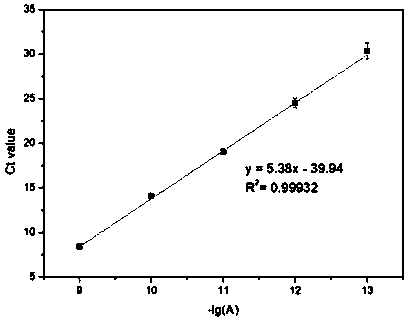
[0062] (3) Real-time fluorescent quantitative PCR: the reaction is carried out on a 96PCR plate, and the reverse transcription product diluted 10 times, 250nM primer II (sequence shown in SEQ ID No.2), 250nM primer III (sequence As shown in SEQ ID No.3) and the reaction system of 1×SYBR Premix ExTMTaqⅡ, then put the plate into 480InstrumentII for PCR reaction. The procedure is to heat at 9...
Embodiment 2
[0065] (1) TdT extension step: first prepare 2 , 10pM DNA-122, 1×TdT reaction buffer and 10U TdT enzyme reaction system, then react at a constant temperature of 37°C for 30min, and then inactivate at 85°C for 25min.
[0066] (2) Reverse transcription step: Prepare a reaction system containing 10-fold diluted reaction product from the previous step, 100nM primer I, 1× reverse transcription buffer and reverse transcriptase MixI, then react at a constant temperature of 37°C for 15min, then 85°C Inactivate 5s.
[0067] (3) Real-time fluorescent quantitative PCR: The reaction was carried out on a 96PCR plate. Firstly, a reaction system containing 10-fold diluted reverse transcription product, 250nM primer II, 250nM primer III and 1×SYBR Premix ExTMTaqII was prepared in the well, and then the plate was put into a 480Instrument II for PCR reactions. The procedure is to heat at 95°C for 30s, then cycle through heating at 95°C for 5s and extension at 63°C for 30s, and terminate the...
Embodiment 3
[0070] (1) TdT extension step: first prepare 2 , 10pM DNA-122, 1×TdT reaction buffer and 10U TdT enzyme reaction system, then react at a constant temperature of 37°C for 30min, and then inactivate at 85°C for 25min.
[0071] (2) Reverse transcription step: Prepare a reaction system containing 10-fold diluted reaction product from the previous step, 100nM primer I, 1× reverse transcription buffer and reverse transcriptase MixI, then react at a constant temperature of 37°C for 15min, then 85°C Inactivate 5s.
[0072](3) Real-time fluorescence quantitative PCR: The reaction was carried out on a 96PCR plate. Firstly, a reaction system containing 10-fold diluted reverse transcription product, 250nM primer II, 250nM primer III and 1×SYBR Premix ExTMTaqII was prepared in the well, and then the plate was put into a 480Instrument II for PCR reactions. The procedure is to heat at 95°C for 30s, then cycle through heating at 95°C for 5s and extension at 63°C for 30s, and terminate the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com