Cancer disease gene characteristic selection method based on historical data
A technology of genetic characteristics and historical data, applied in the fields of instruments, character and pattern recognition, biostatistics, etc., can solve the problems of poor practicability, high time complexity, and many redundant features, so as to improve the accuracy of prediction, The effect of reducing data dimensionality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0038] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with specific embodiments and with reference to the accompanying drawings.
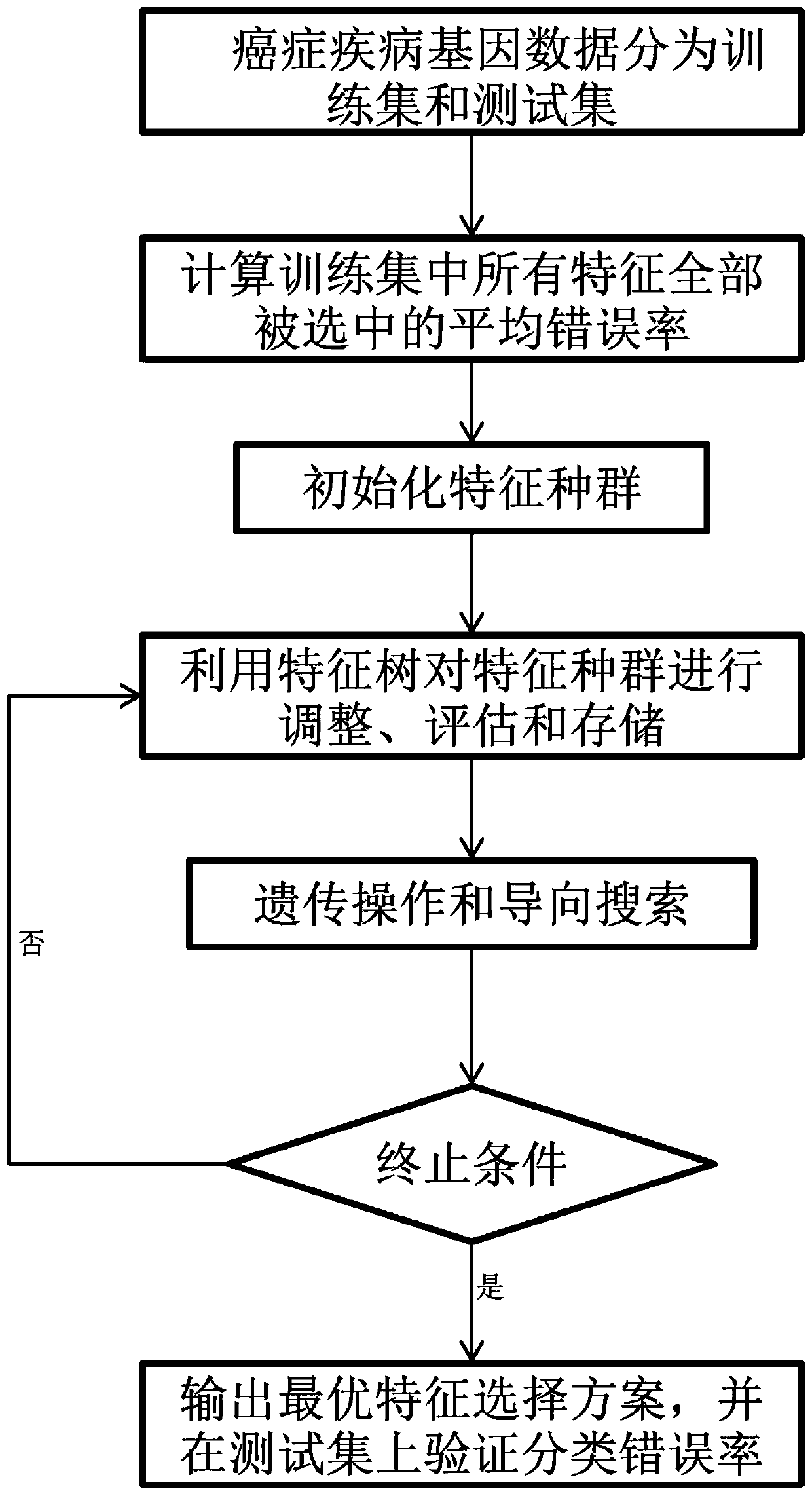
[0039] Such as figure 1 As shown, this embodiment provides a method for selecting gene features of cancer diseases based on historical data, including the following steps:
[0040] Step A: divide the cancer disease gene data into a training data set and a test data set; specifically, divide the cancer disease gene data into ten parts, 7 parts are used as training data sets, and 3 parts are used as test data sets.
[0041] Step B: Calculate the total average error rate after all the features on the training data set are selected by using the 50-fold crossover, specifically: divide the training data set into five parts on average, select one as the test set, and the other four as the training set get error rate where CE ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com