Integrated purification and measurement of DNA methylation and co-measurement of mutations and/or mRNA expression levels in an automated reaction cartridge
A technology of methylation and cassette group, which is applied in the direction of DNA preparation, recombinant DNA technology, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
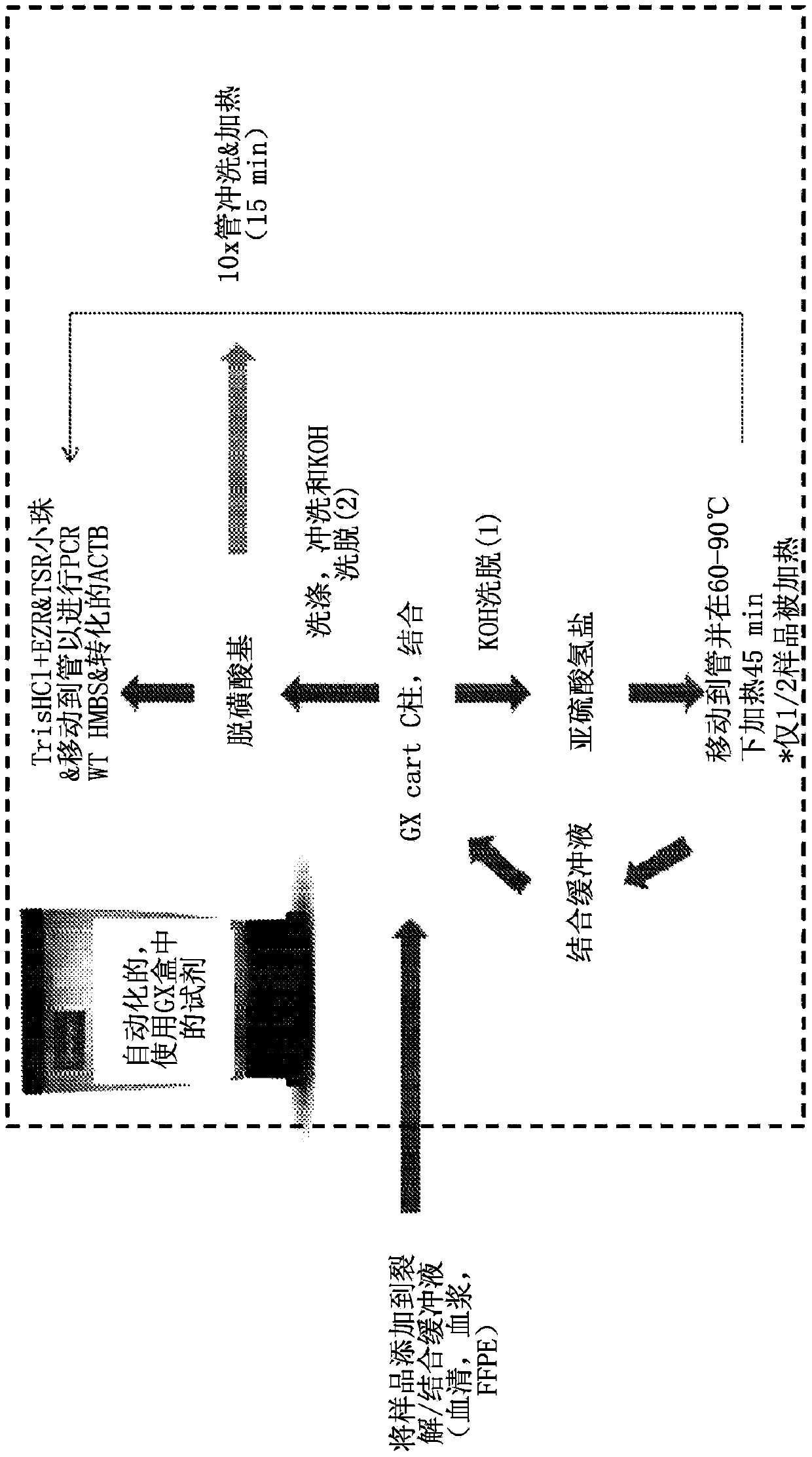
[0997] To validate the method, human genomic DNA (HGDNA) was used as a starting sample to monitor Cepheid In-box sample preparation, bisulfite conversion, sample cleanup, and methylation-specific qPCR. To measure bisulfite conversion efficiency, half of the DNA-bisulfite mixture was loaded into 50 µL cassette tubes during the bisulfite conversion step and heated. Therefore, under optimal transformation conditions, about half of the HGDNA is transformed and the other half remains untransformed.
[0998] Primers and Taqman probes for the qPCR step were designed for a non-transformed gene (HMBS (hydroxymethylbilane synthase housekeeping gene)) and a transformed gene (ACTB (beta-actin)), then compared The cycle threshold (Ct) was used to quantify the conversion efficiency. Both ACTB and HMBS are commonly used as single-copy or low-copy reference genes, so the inventors expected similar copy numbers per ng of HGDNA.
[0999] Representative of runs from 300ng HGDNA shown below...
Embodiment 2
[1001] Figure 6A and 6B Linearity of transformed ACTB is shown. in particular, Figure 6A Results of 15 cycles of nested qPCR using hgDNA, ACTB are shown. As can be seen from the right panel, the signal (Ct value) is essentially linear between about 25,000 copies and about 100 copies. Figure 6B Results of 20 cycles of nested qPCR using hgDNA, ACTB are shown. These figures demonstrate the sensitivity of the cassette to hgDNA. Dropout begins at about 20-50 copies with a sensitivity of about 25 copies of transforming DNA.
[1002] Figure 7A , 7B and 7C show qPCR results for six methylated targets (AKR1B1, HOXB4, TM6SF1, RASGRF2 and RASSF1A). Figure 7A Nested qPCR of 20 cycles of control (25 ng HSDNA and 5000 MBA-453 cells whose DNA was not converted by bisulfite) is shown. Figure 7B Shown are the results of 20 cycles of nested PCR of six methylated targets using DNA from MBA-453 cells that had been transformed with bisulfite. Strong signals for all targets are show...
Embodiment 3
[1004] Figure 8 The results of the measurement of transformation efficiency are illustrated. The difference in conversion efficiency between unconverted HMBS and transformed ACTB was about 66% (~1 Ct). The ideal Ct for 100% bind / elute, 100% conversion and 100% bind-elute is about 24-25. For a 10-fold reduction, experiments appear to show 50% bind / elute, 50-66% conversion and 50% bind / elute with a Ct of approximately 27.
[1005] Figure 9 The increased specificity of transformed DNA generated by nested qPCR is illustrated. Nested PCR appears to increase the specificity of transformed DNA, increase the specificity of methylated DNA, and reduce contamination problems.
[1006] Figure 10 The specificity of the methylation cassette is illustrated. Specificity is not shown for untransformed DNA (upper panel) or unmethylated DNA (lower panel), except for HIST1H3C.
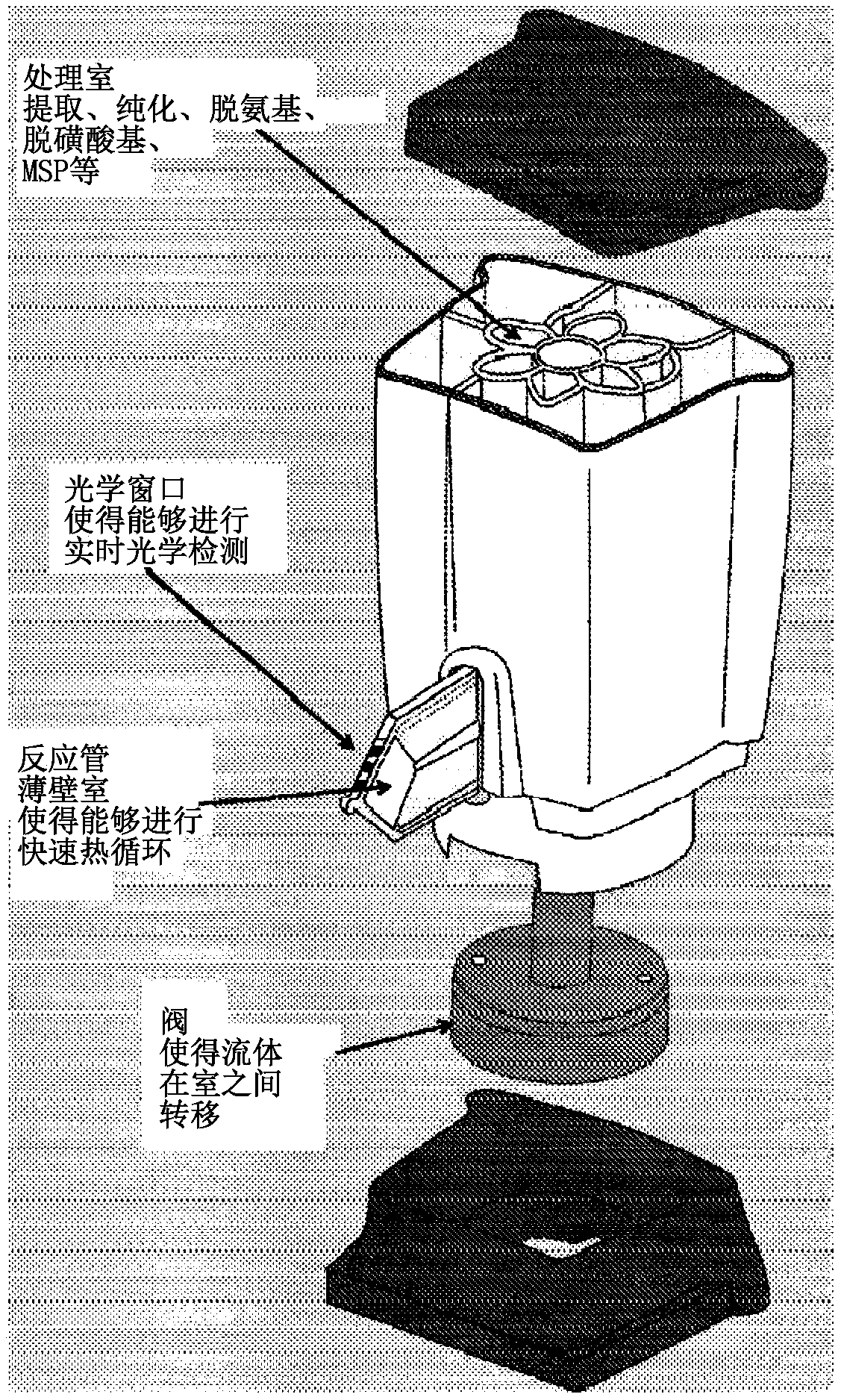
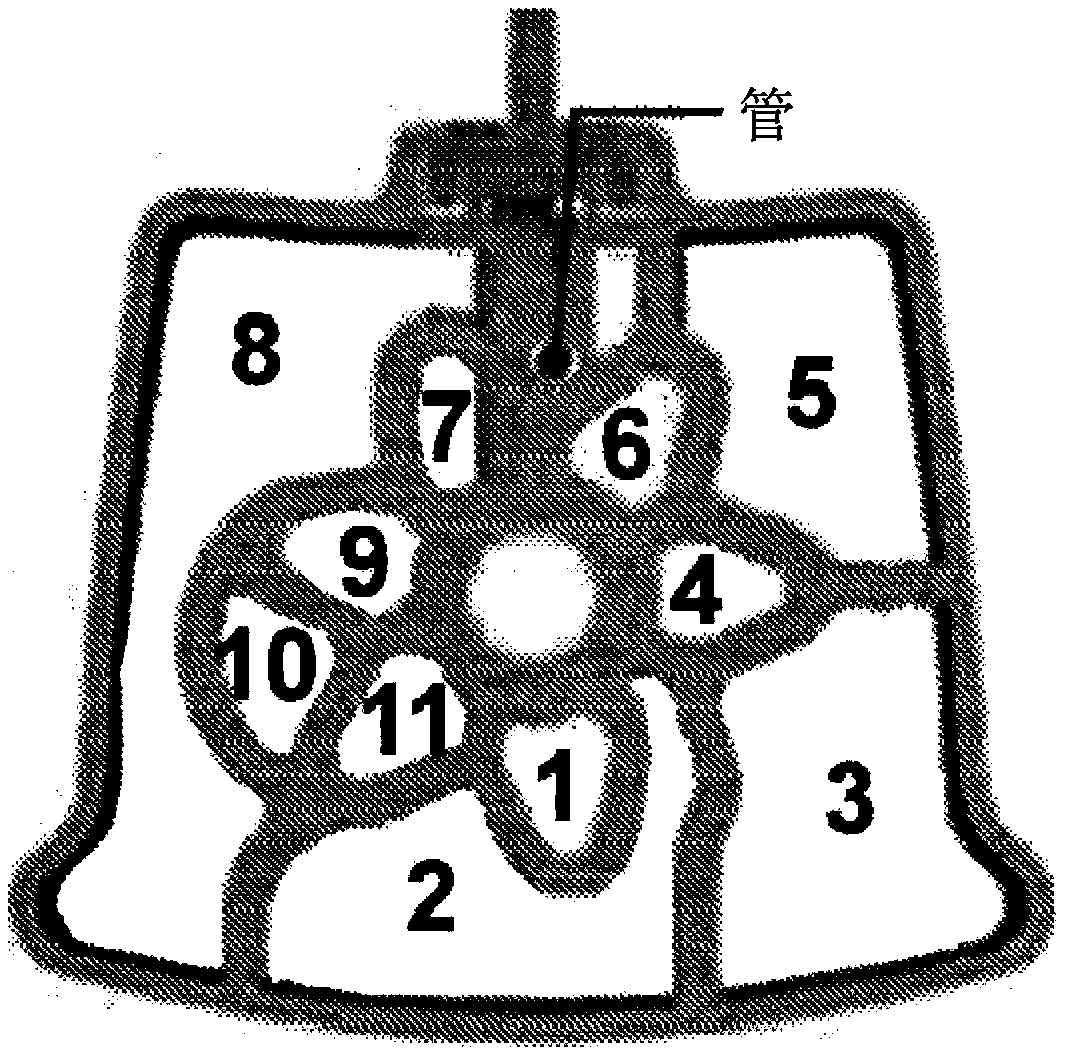
[1007] Figure 11 A and 11B show the use of cartridges (e.g. box) for some illustrative but non-specific f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com