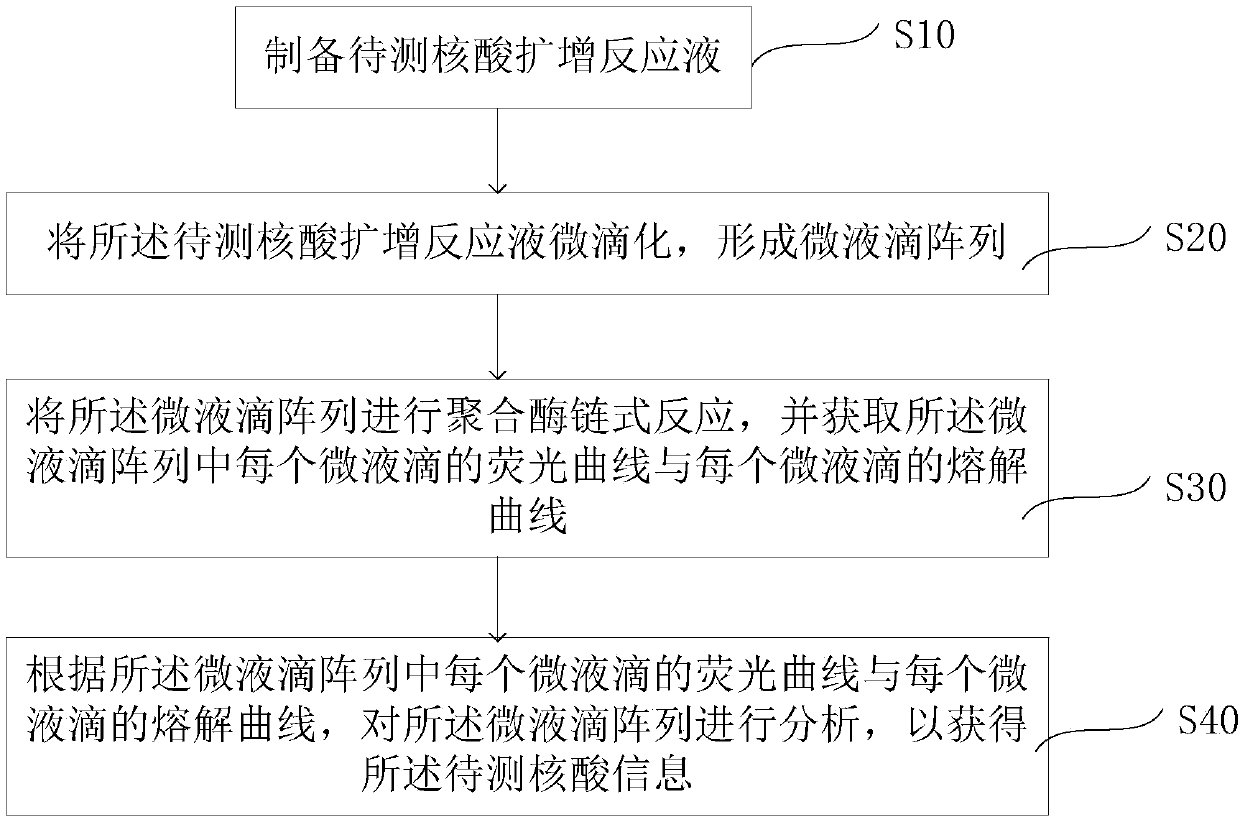
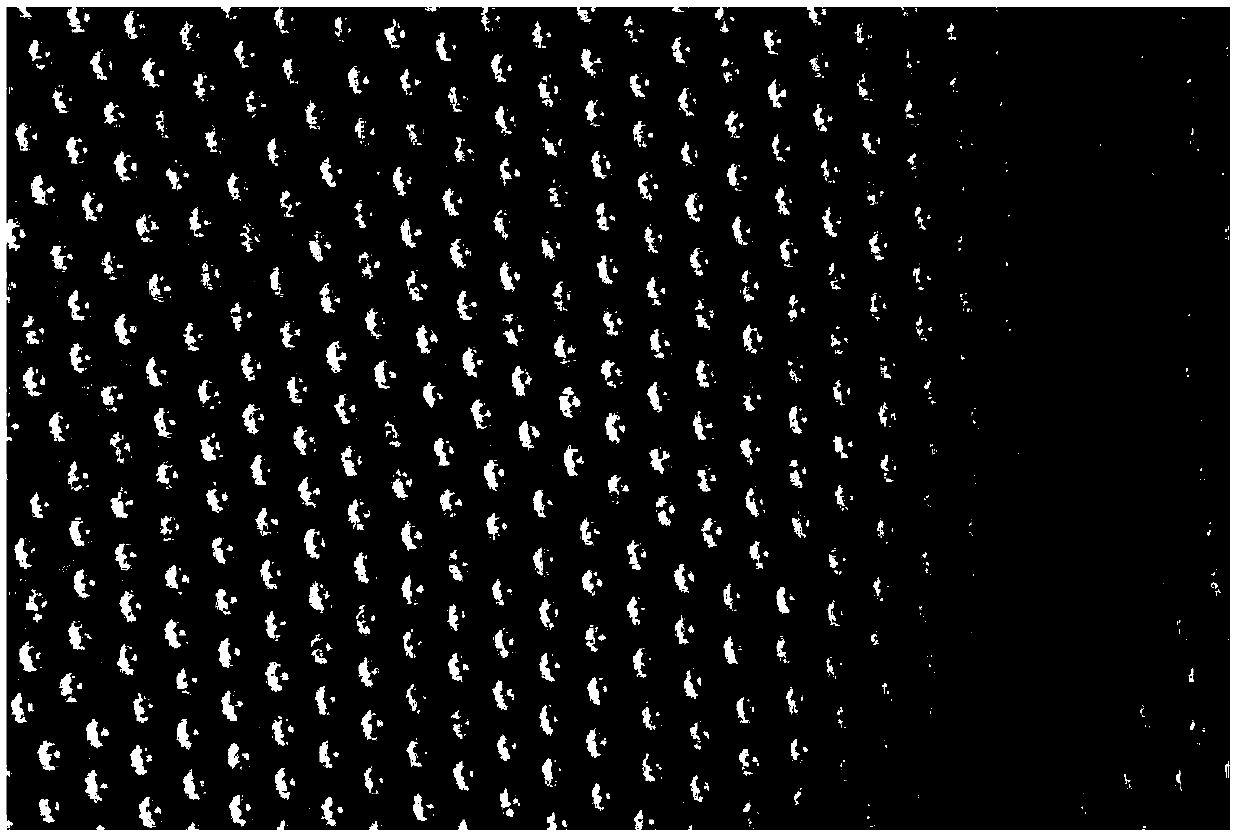
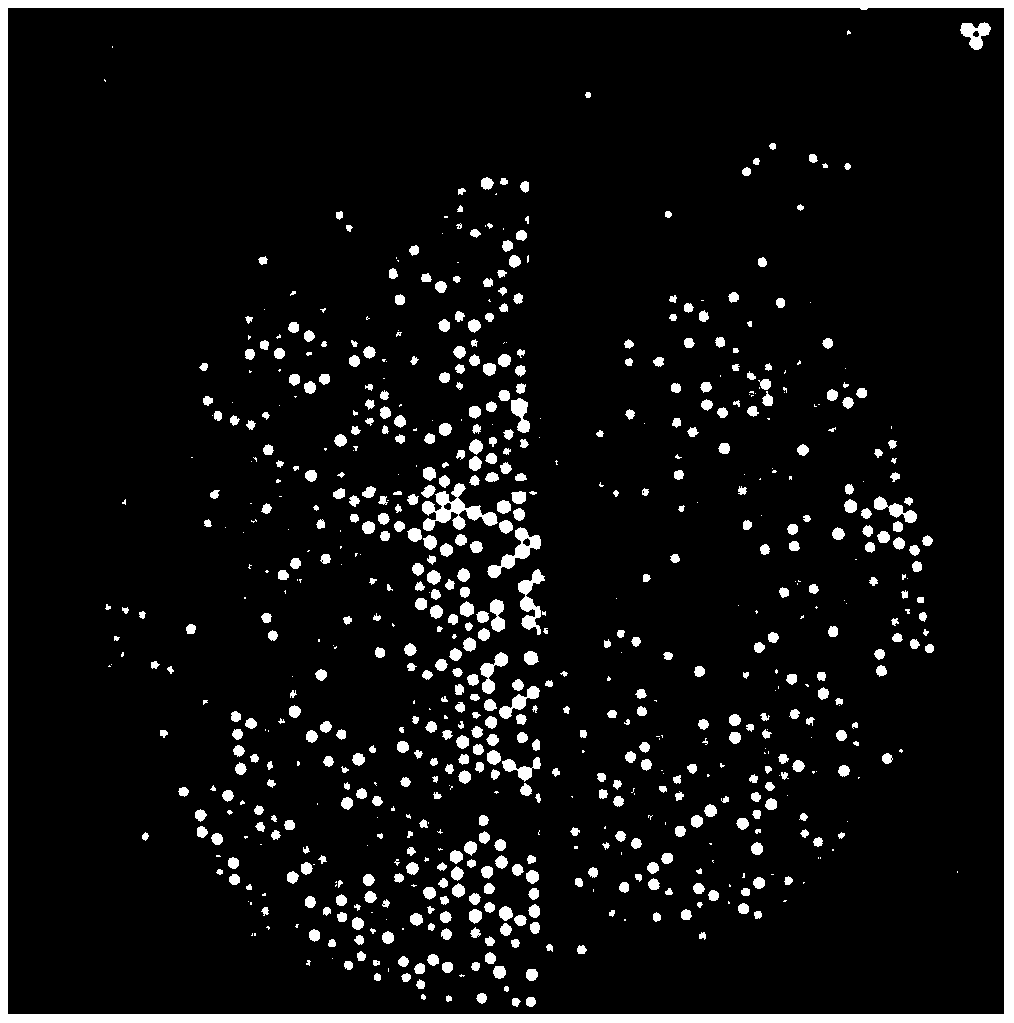
Patents
Literature
69 results about "High Resolution Melt Analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
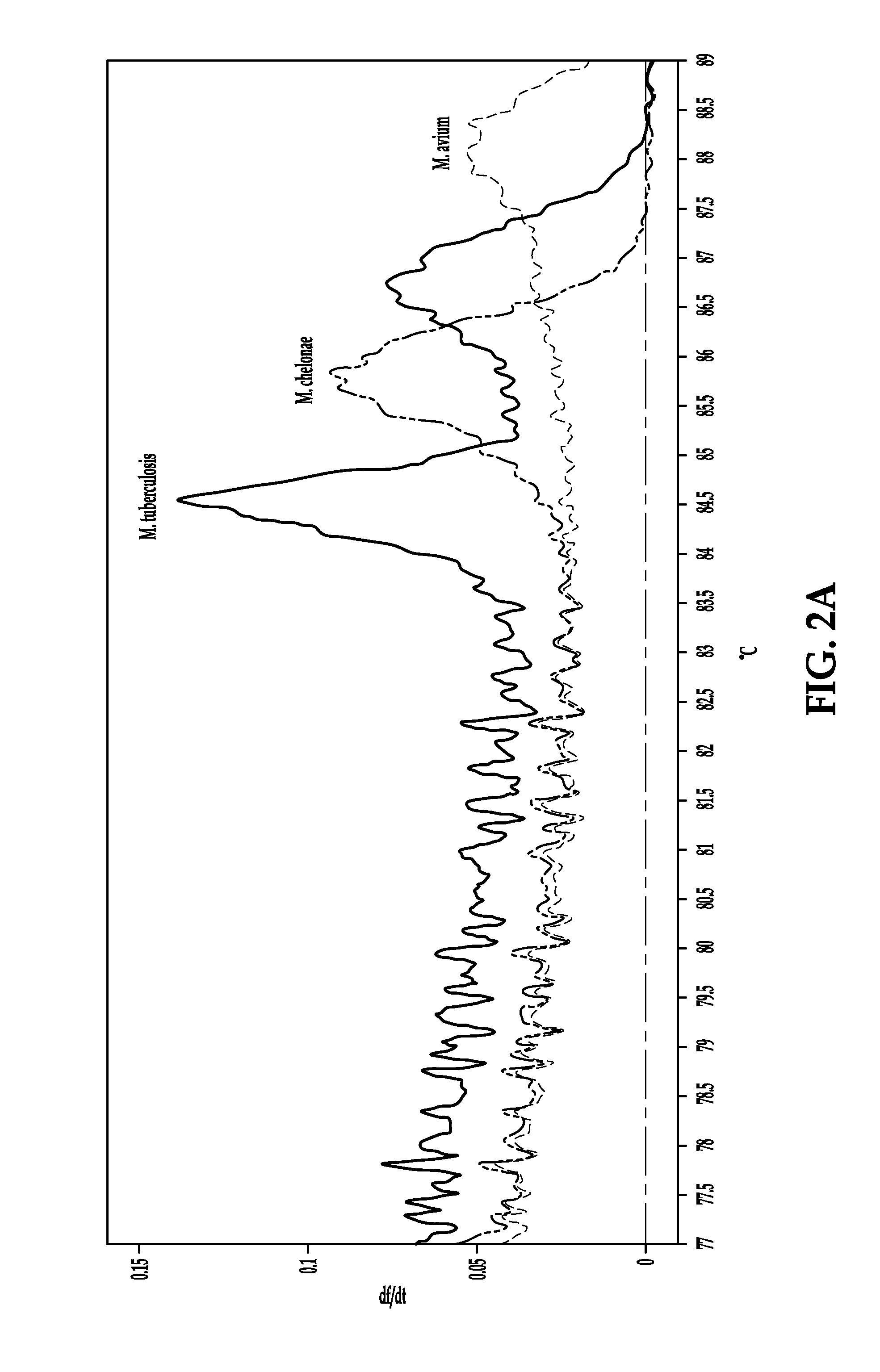
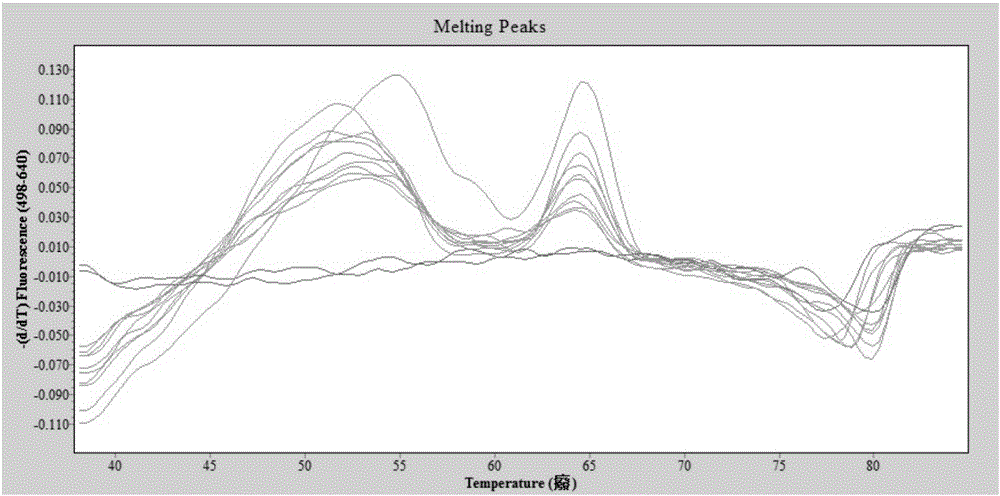
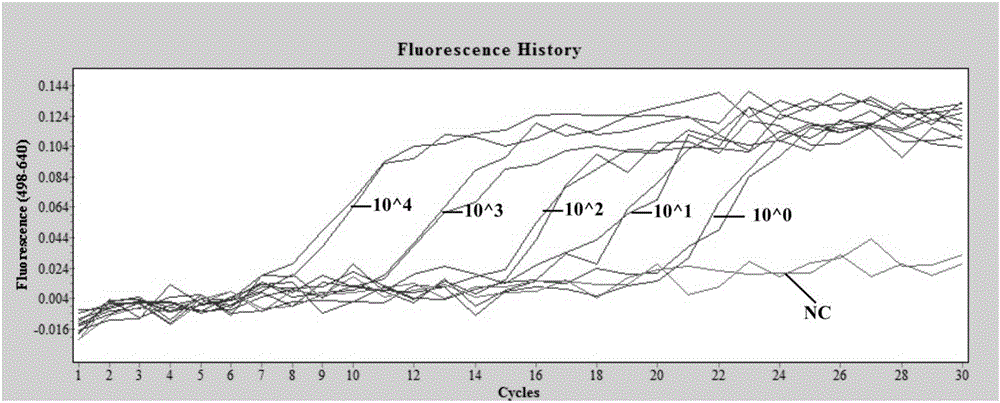
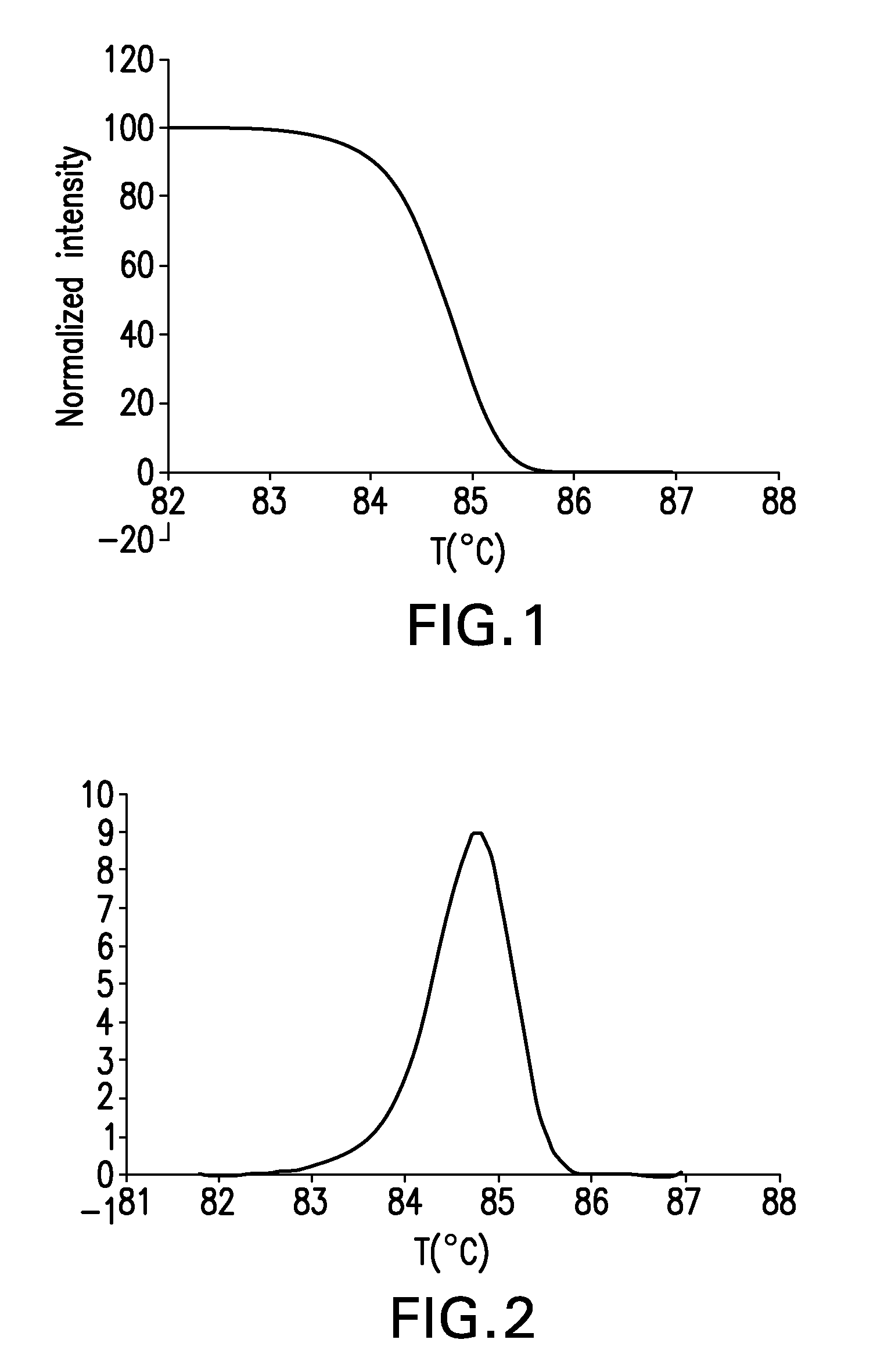
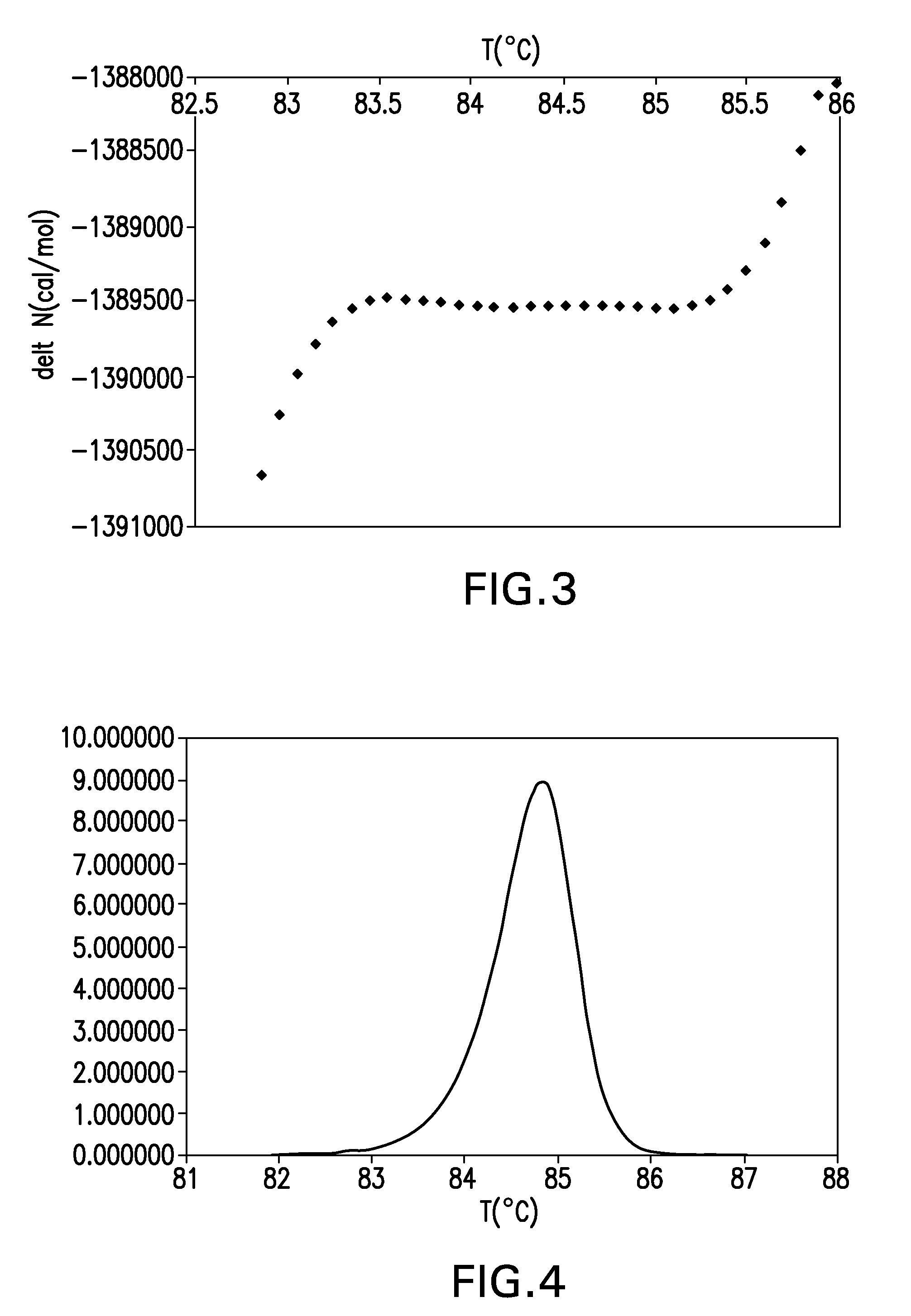
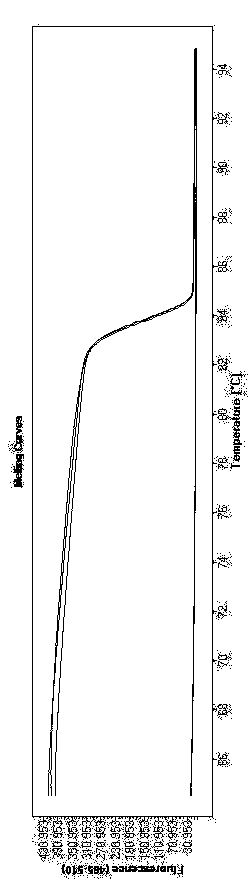
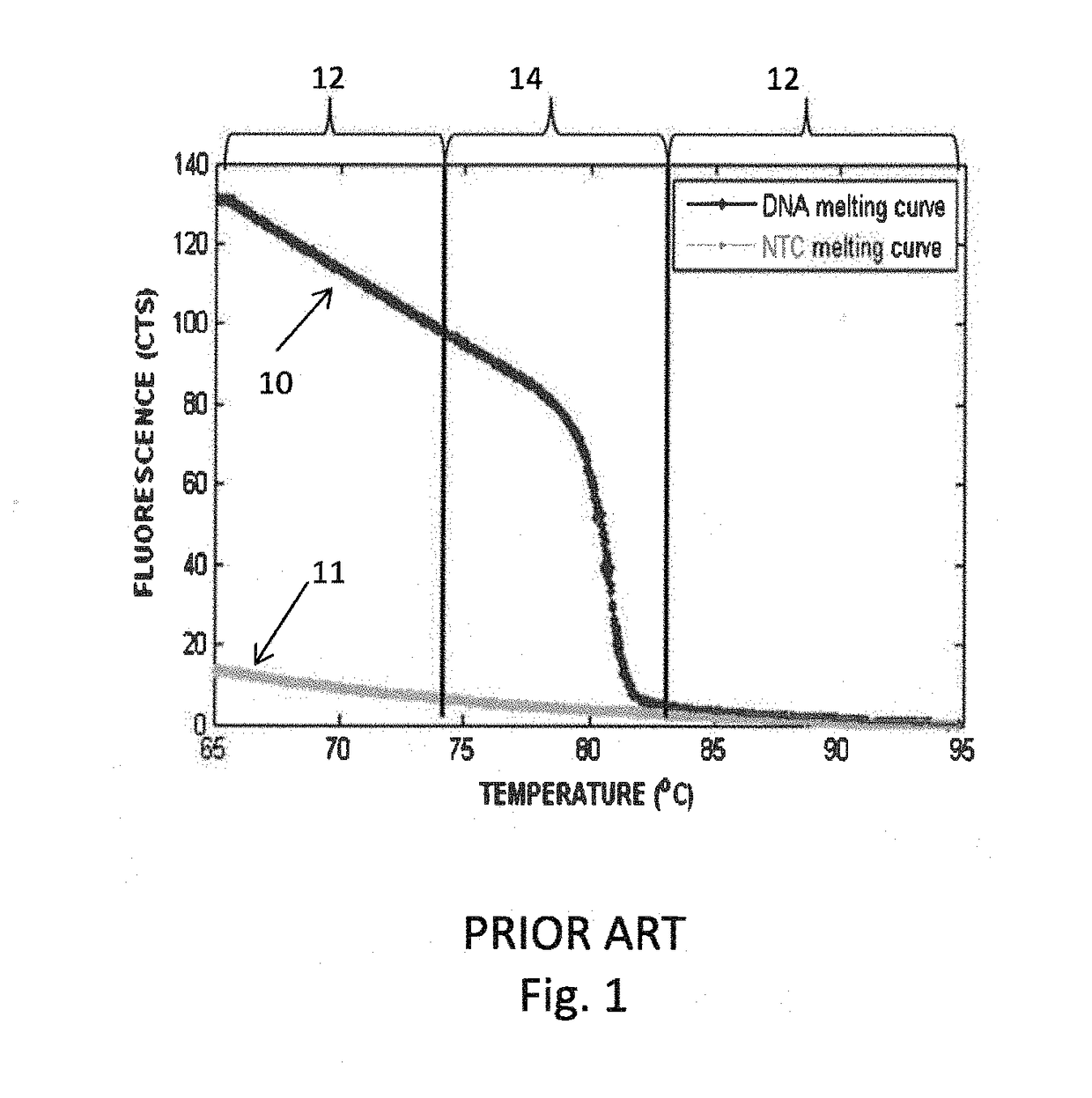
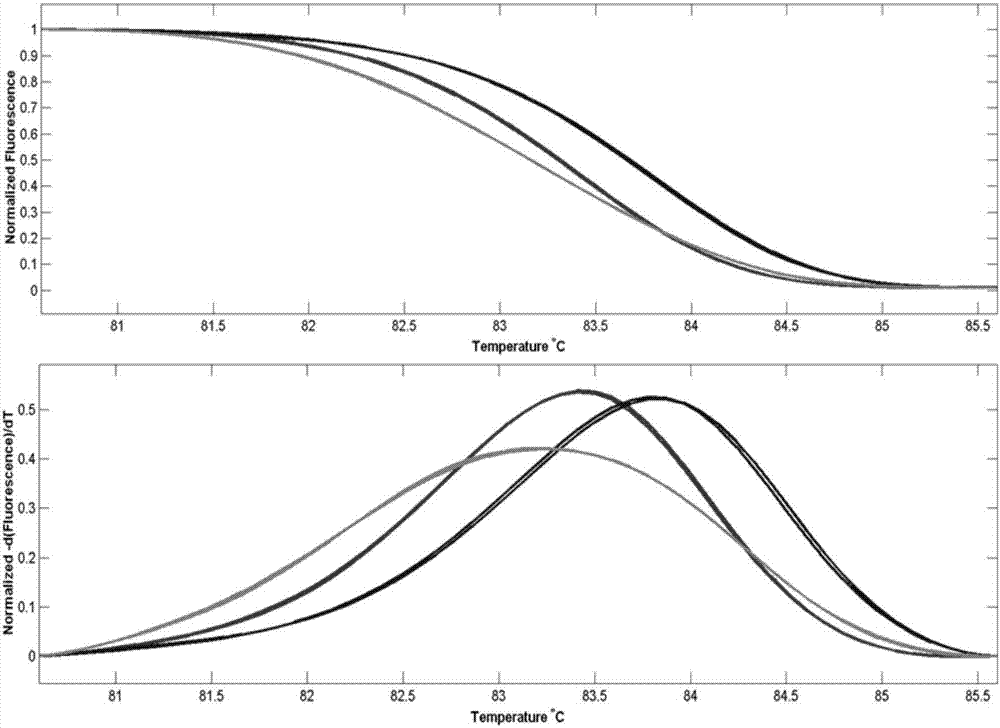
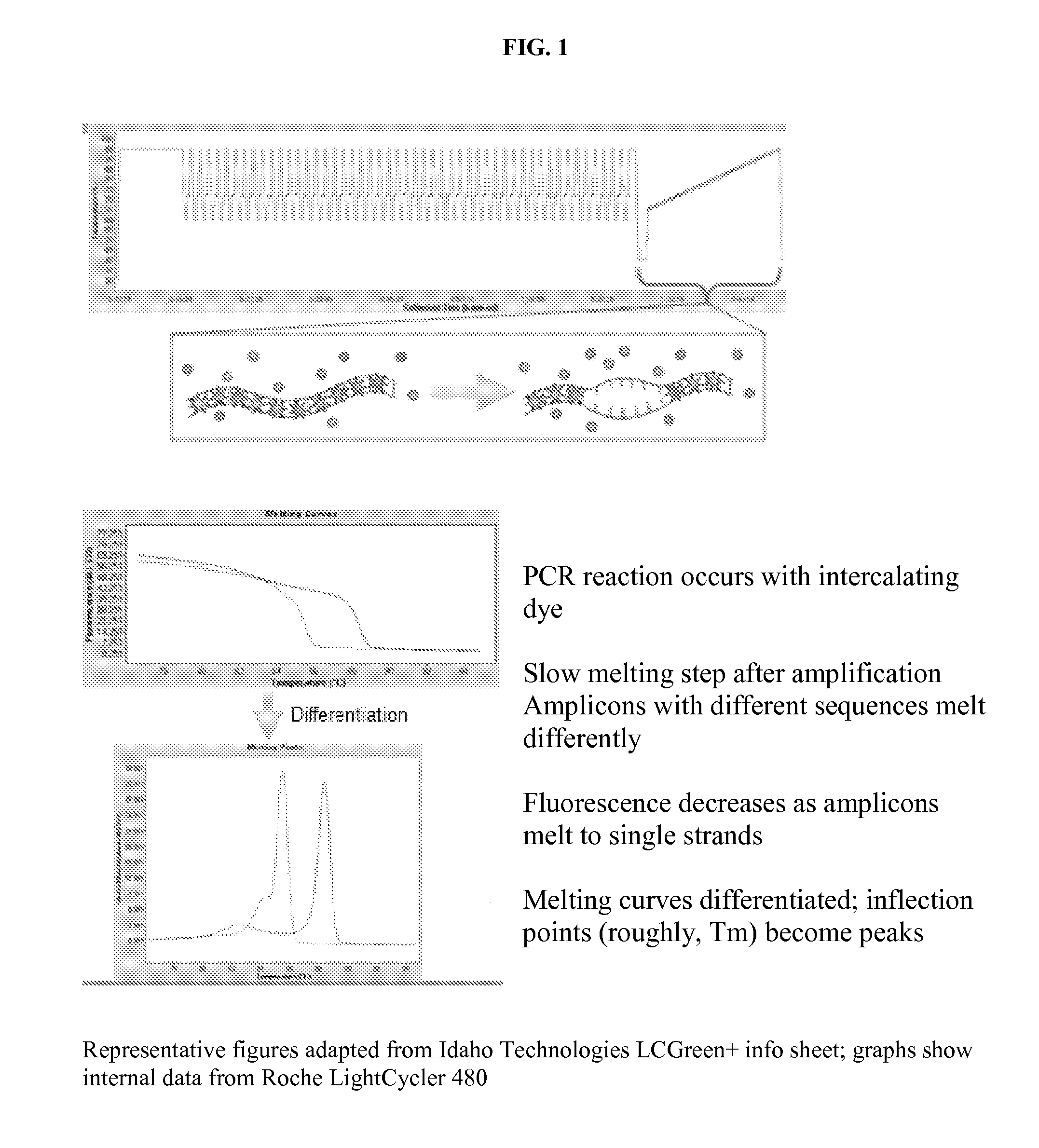
A reverse, real-time, polymerase chain reaction technique in which fluroescently labeled double-stranded DNA is slowly melted apart by increasing temperature. Genetic mutations and polymorphisms can be detected by analyzing the melt curves, which correspond to the rate at which double-stranded DNA become single-stranded.
Preparation method of non-transgenic CRISPR mutant
InactiveCN108611364AReduce frequencyImprove accuracyNucleic acid vectorVector-based foreign material introductionScreening methodSexual reproduction
The invention provides a preparation method of a non-transgenic mutant based on a CRISPR-Cas9 technology. Specifically, the preparation method comprises a construction method and a screening method. The construction method is characterized in that CRISPR-Cas9 and sgRNA are preassembled and are located at T-DNA of agrobacterium tumefaciens; site-directed change of a target gene of a target plant can be realized by infection of the target plant by the agrobacterium tumefaciens and coculture without integrating the sequences of the CRISPR-Cas9 and the sgRNA into a genome of the target plant, so that an obtained site-directed mutant material of the target gene does not need the processes of sexual reproduction, segregation posterity, population screening and the like or does not contain any exogenous gene sequence. The obtained regeneration seedlings are screened by a high-throughput sequencing and high-resolution melting curve technology provided by the invention; mutant plants of which target genes are mutated can be obtained by identifying the regeneration seedlings efficiently and quickly at the current generation of transgene, even if low-proportion mutants of which the proportionis 1 / 100 of a mixed population can be screened. The screening method provided by the invention still can ensure excellent accuracy and sensitivity.
Owner:NANJING AGRICULTURAL UNIVERSITY
Method, database and primer pair for identifying herbal medicinal materials
The invention provides a method for identifying types of herbal medicinal materials. The method comprises the following steps: obtaining a DNA sample of a herbal medicinal material to be detected; (b) performing a polymerase chain reaction between the DNA sample of the herbal medicinal material to be detected and at least one primer pair to obtain a polymerase chain reaction product; (c) performing high-resolution melting analysis on the polymerase chain reaction product to obtain a high-resolution melting analysis spectrum of the herbal medicinal material to be detected; and (d) comparing the high-resolution melting analysis spectrum of the herbal medicinal material to be detected with a high-resolution melting analysis spectrum of a known herbal medicinal material.
Owner:IND TECH RES INST
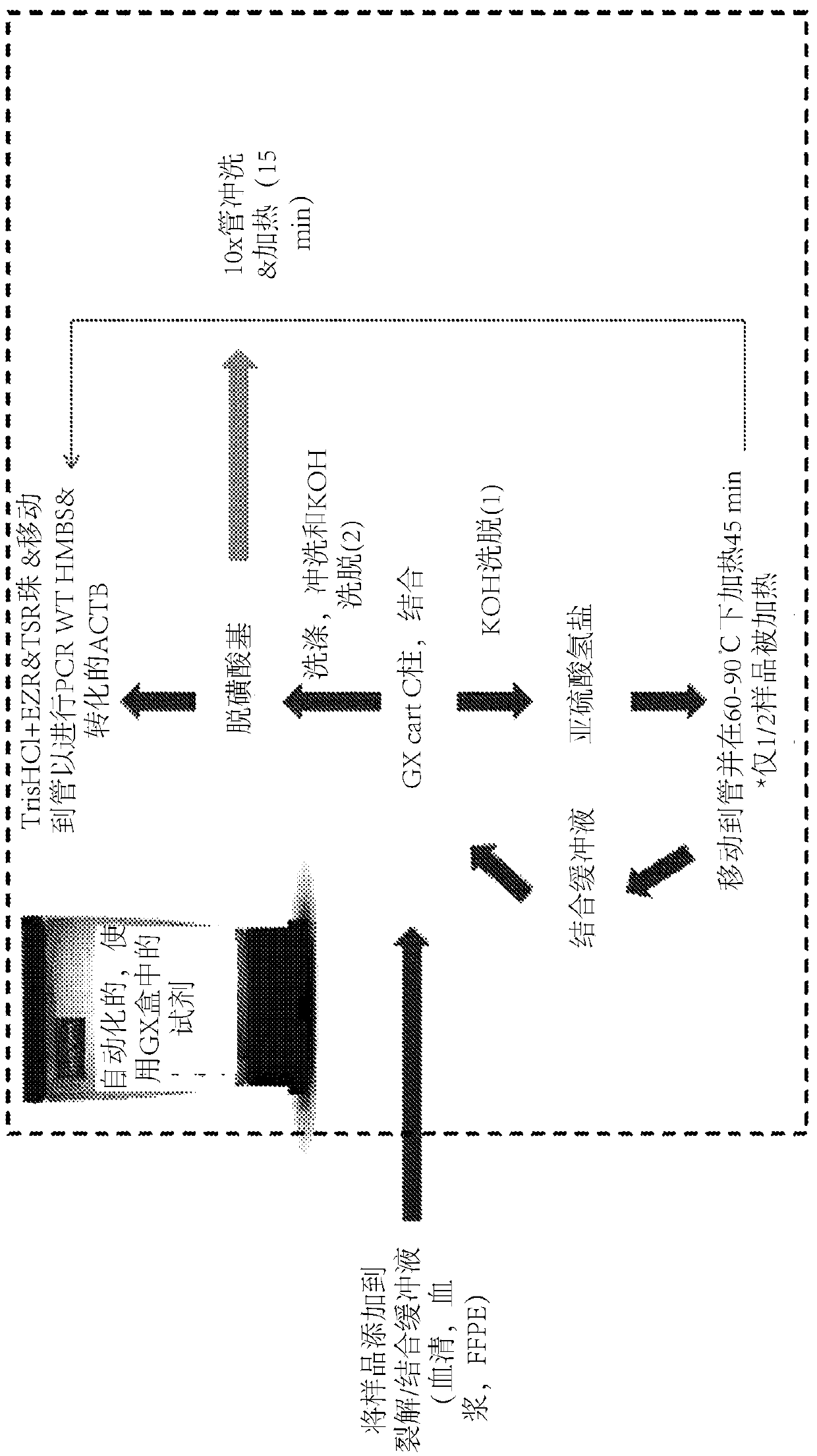
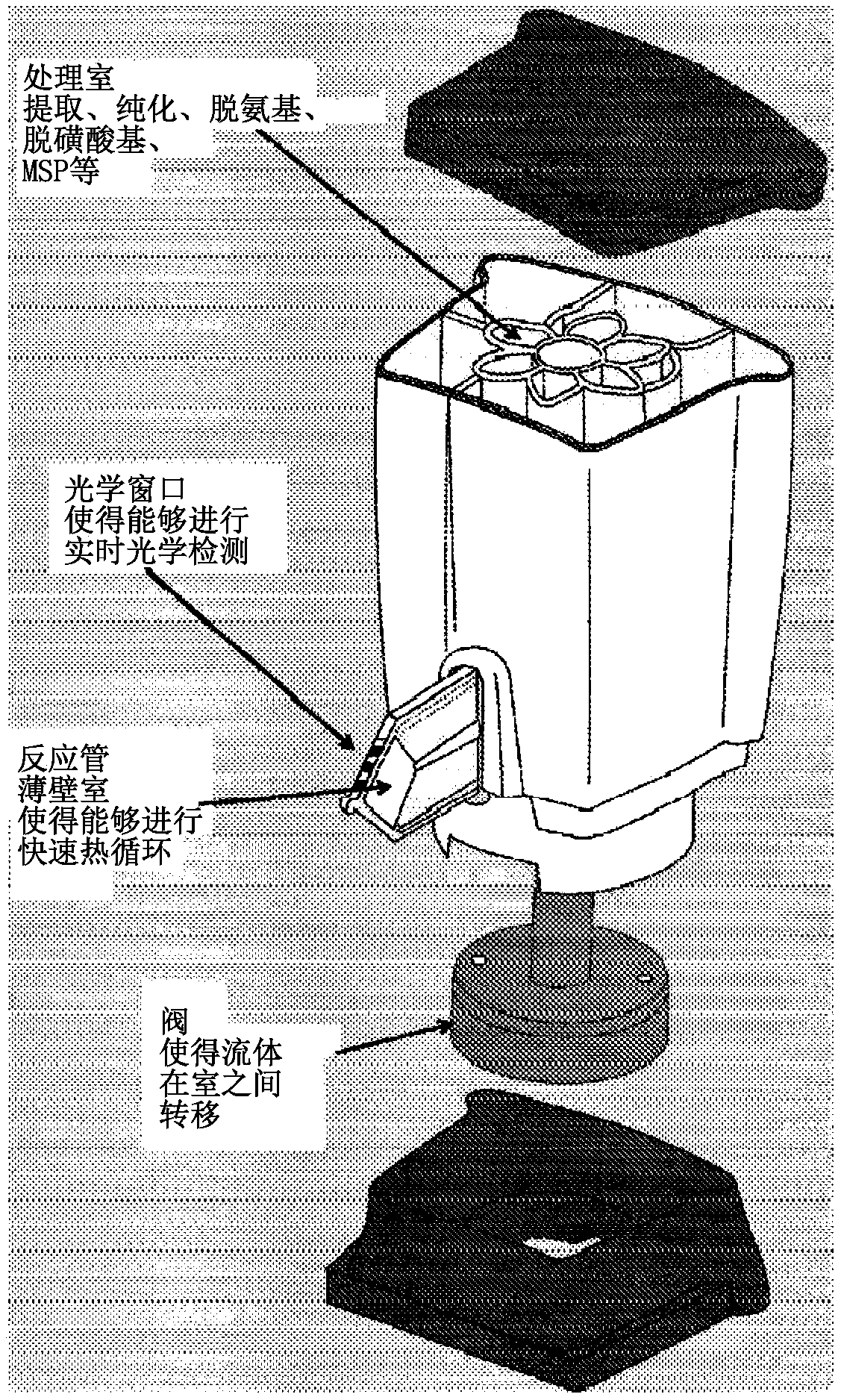
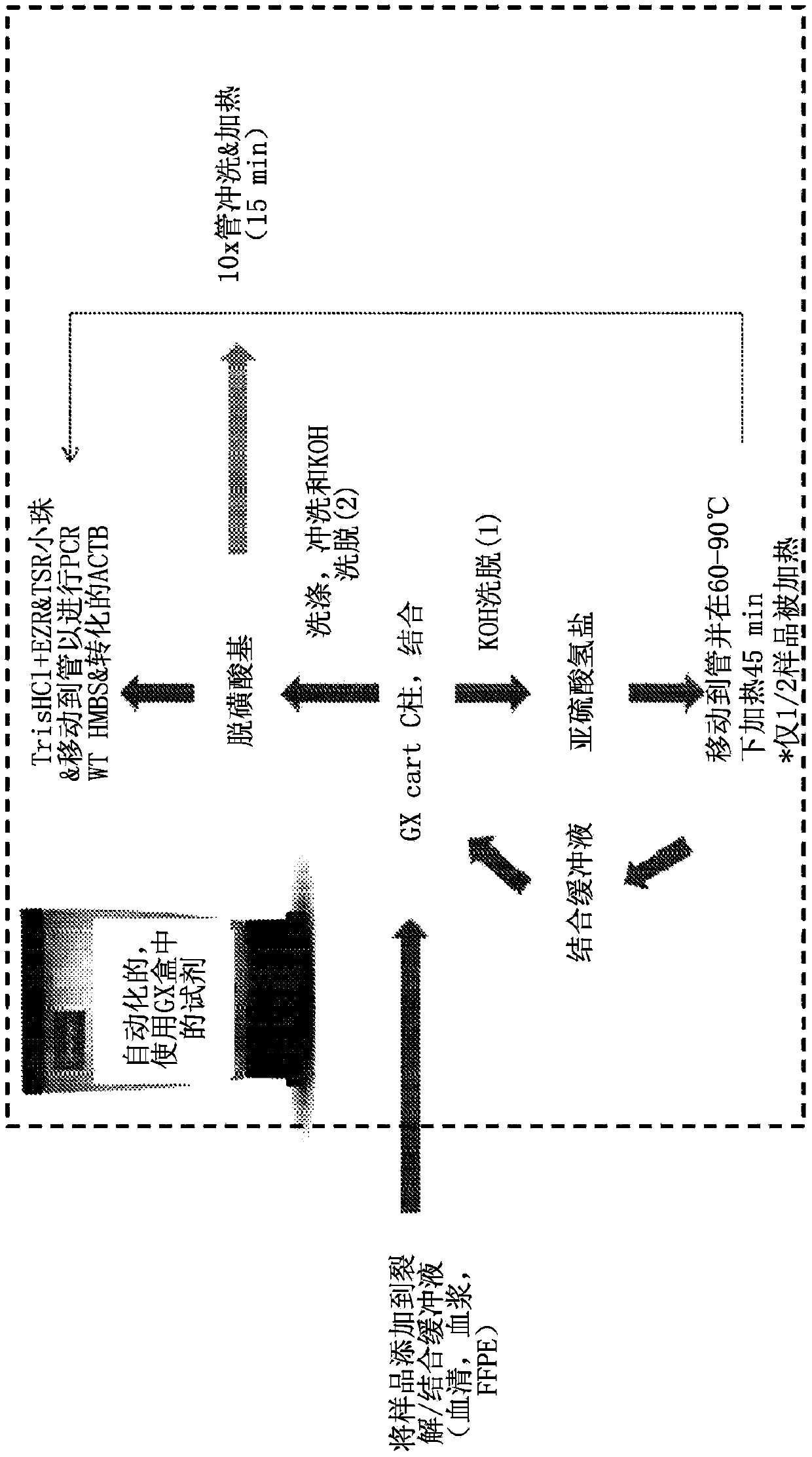
Integrated purification and measurement of dna methylation and co-measurement of mutations and/or mrna expression levels in automated reaction cartridge
PendingCN107922941AHeating or cooling apparatusMicrobiological testing/measurementNucleic acid sequencingBisulfite
In various embodiments methods of determining methylation of DNA are provided. In one illustrative, but non-limiting embodiment the method comprises i) contacting a biological sample comprising a nucleic acid to a first matrix material comprising a first column or filter where said matrix material binds and / or filters nucleic acids in said sample and thereby purifies the DNA; ii) eluting the boundDNA from the first matrix material and denaturing the DNA to produce eluted denatured DNA; iii) heating the eluted DNA in the presence of bisulfite ions to produce a deaminated nucleic acid; iv) contacting said deaminated nucleic acid to a second matrix material comprising a second column to bind said deaminated nucleic acid to said second matrix material; v) desulfonating the bound deaminated nucleic acid and / or simultaneously eluting and desulfonating the nucleic acid by contacting the deaminated nucleic acid with an alkaline solution to produce a bisulfite converted nucleic acid; vi) eluting said bisulfite converted nucleic acid from said second matrix material; and vii) performing methylation specific PCR and / or nucleic acid sequencing, and / or high resolution melting analysis (HRM) onsaid bisulfite-converted nucleic acid to determine the methylation of said nucleic acid, wherein at least steps iv) through vi) are performed in a single reaction cartridge.
Owner:CEPHEID INC
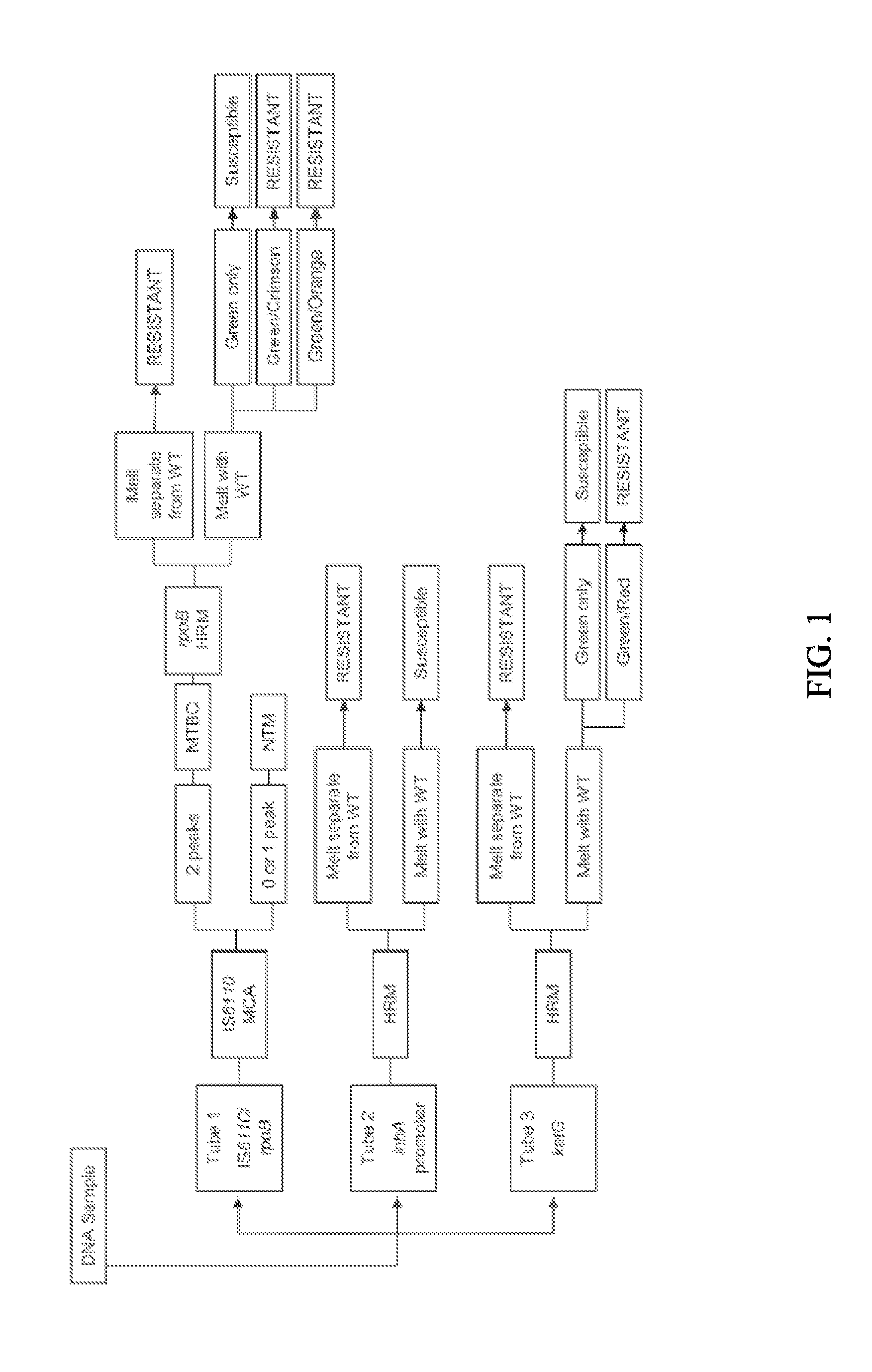
Process for detection of multidrug resistant tuberculosis using real-time PCR and high resolution melt analysis
InactiveUS20130095489A1Easy to understandMicrobiological testing/measurementSpecific detectionResistant tuberculosis
Compositions and process are provided for the rapid and specific detection of drug resistant forms of Mycobacterium tuberculosis based on real time PCR and high resolution melt analysis. The compositions and processes are useful for the detection of mutations within the Rifampicin Resistance Determinant Region (RRDR) of rpoB for the detection of rifampicin (RIF) and within specific regions of katG and the inhA promoter for the detection of isoniazid (INH) resistance. The invention also is capable of rapidly discriminating Mycobacterium tuberculosis complex (MTBC) strains from Nontuberculous Mycobacteria (NTM) strains.
Owner:US DEPT OF HEALTH & HUMAN SERVICES
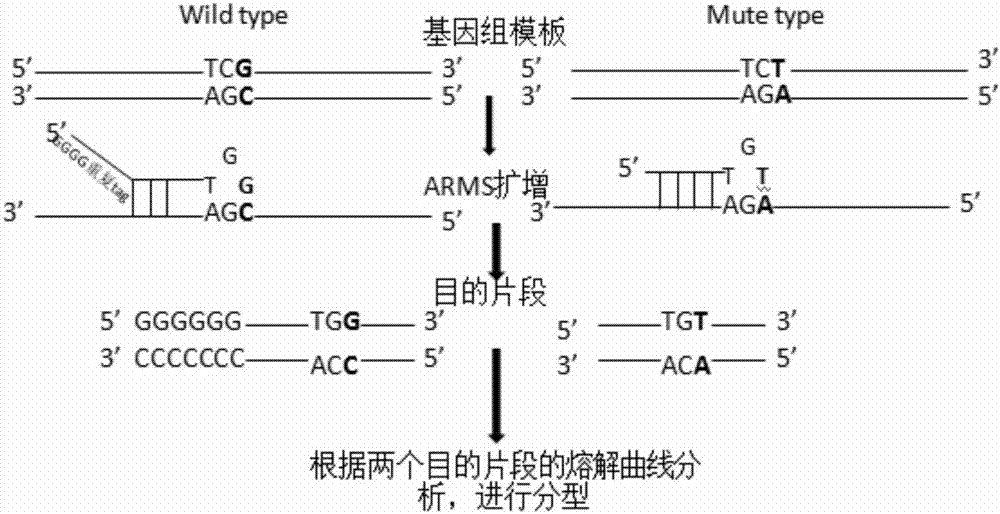
Human ApoE gene polymorphism detection kit based on ARMS-PCR melting curve method
PendingCN107460235AImprove the ability of SNP typingIncrease the difference in melting curvesMicrobiological testing/measurementFluorescenceTyping
The invention provides a human ApoE gene polymorphism detection kit based on the ARMS-PCR melting curve detection method. The human ApoE gene polymorphism detection kit comprises primers for detecting the polymorphism of an ApoE gene, and the primers include two groups of primers for detecting the c.388T>C site and the c.526C>T site respectively. The invention further provides an ARMS-PCR melting curve detection method and a PCR method of the human ApoE gene polymorphism detection kit. By the adoption of the human ApoE gene polymorphism detection kit based on the ARMS-PCR melting curve method, the ability of allele-specific primers to distinguish different alleles can be greatly improved, the difference between melting curves of two amplified allele fragments is greatly increased, the ability of SNP typing of the melting curve method is enhanced, melting curve method SNP typing can be achieved simply with an ordinary fluorescence PCR instrument instead of a high-precision high-resolution melting curve PCR instrument, the detection result is accurate, sensitivity is high, and accurate typing of a genomic DNA sample as low as 1 ng can be achieved.
Owner:SHANGHAI BIOMED UNION BIOTECHNOLOGY CO LTD
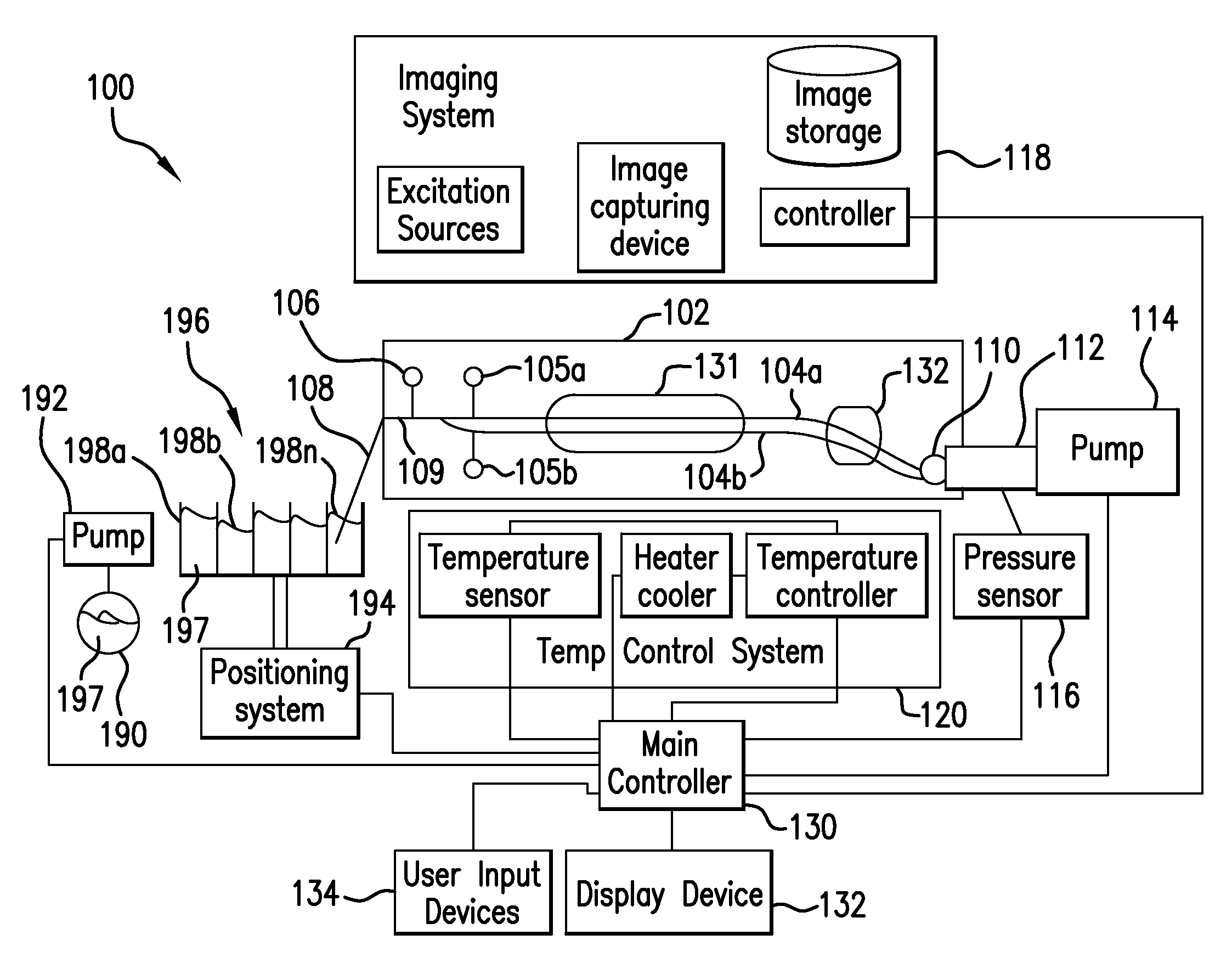
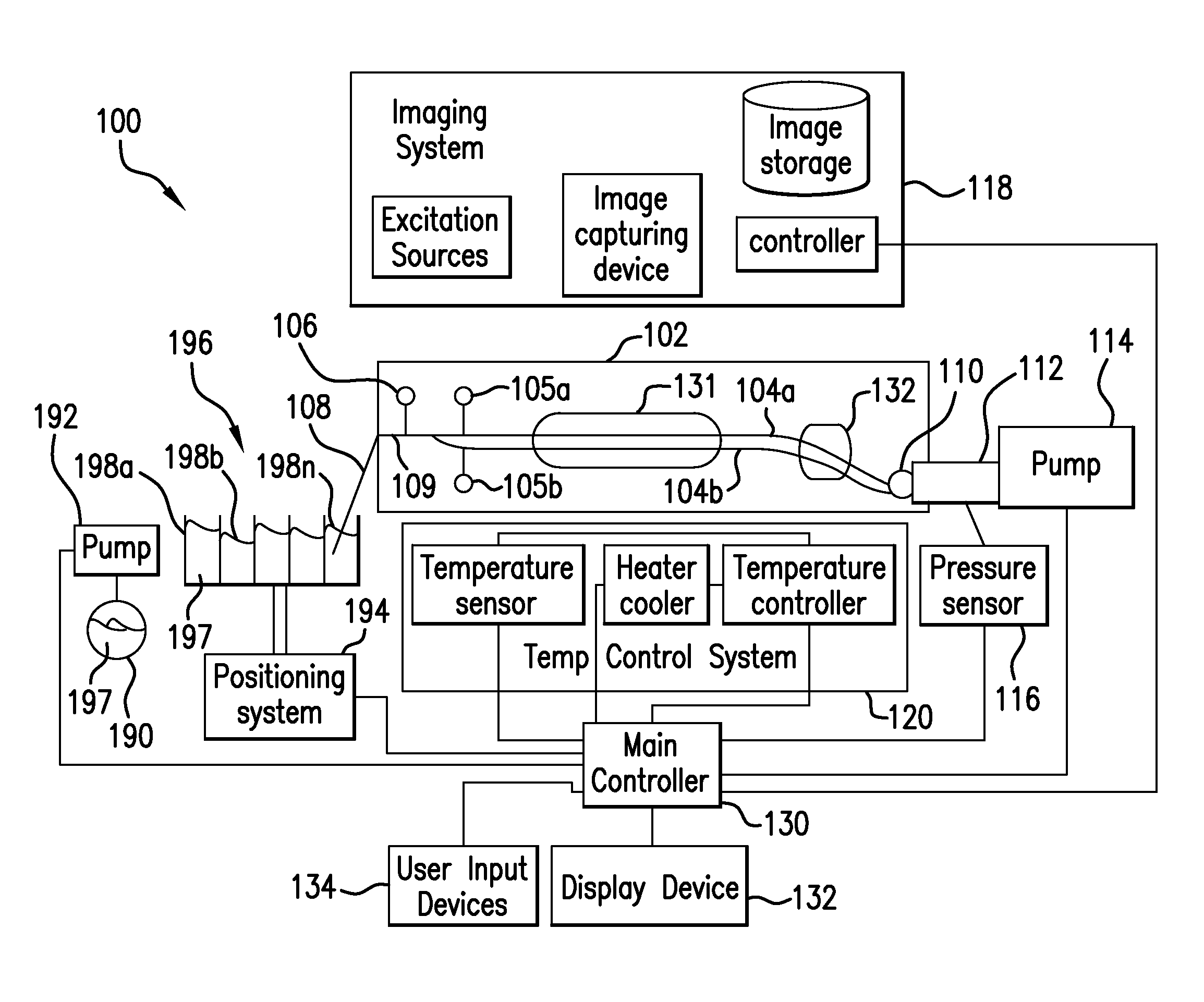
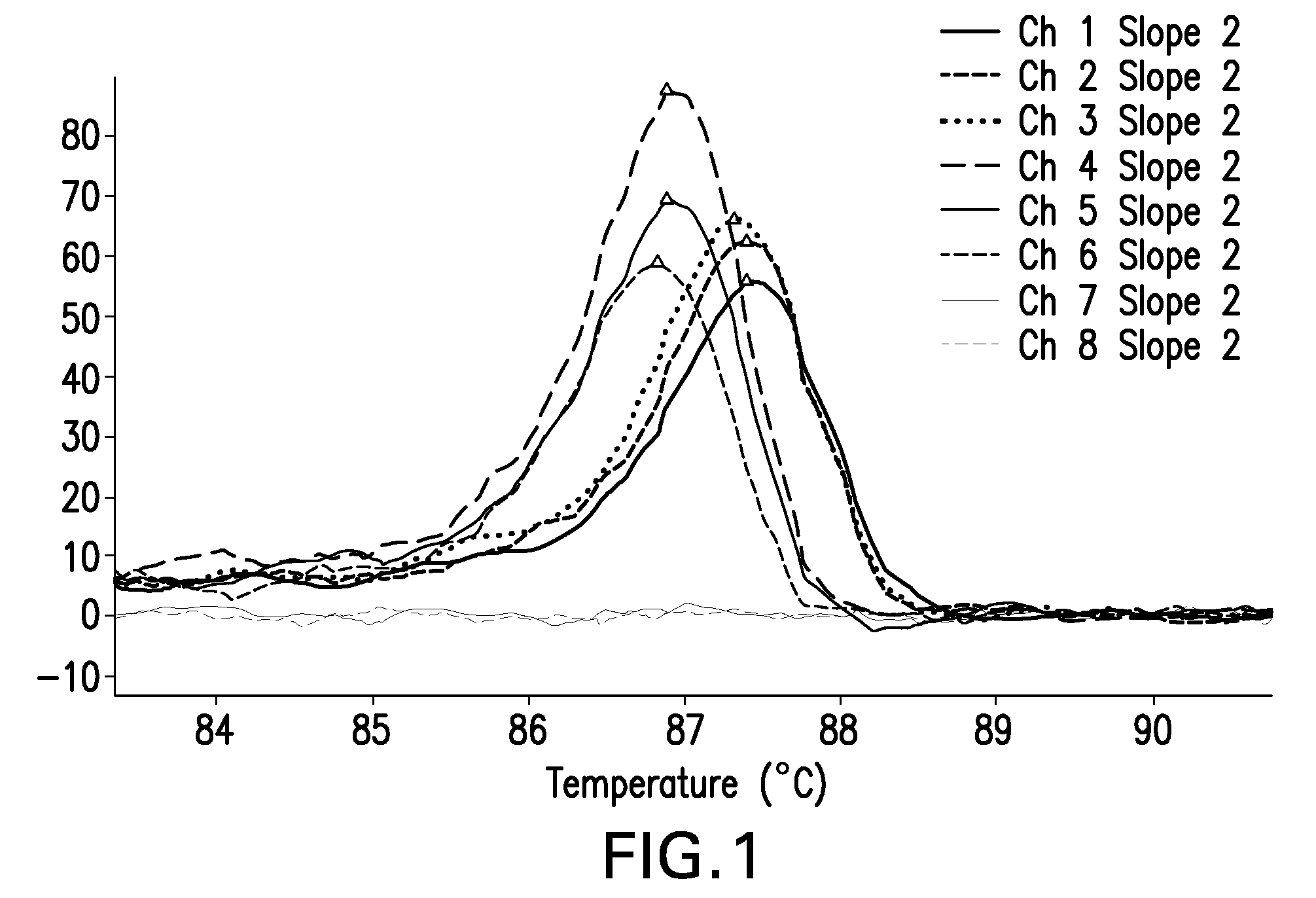
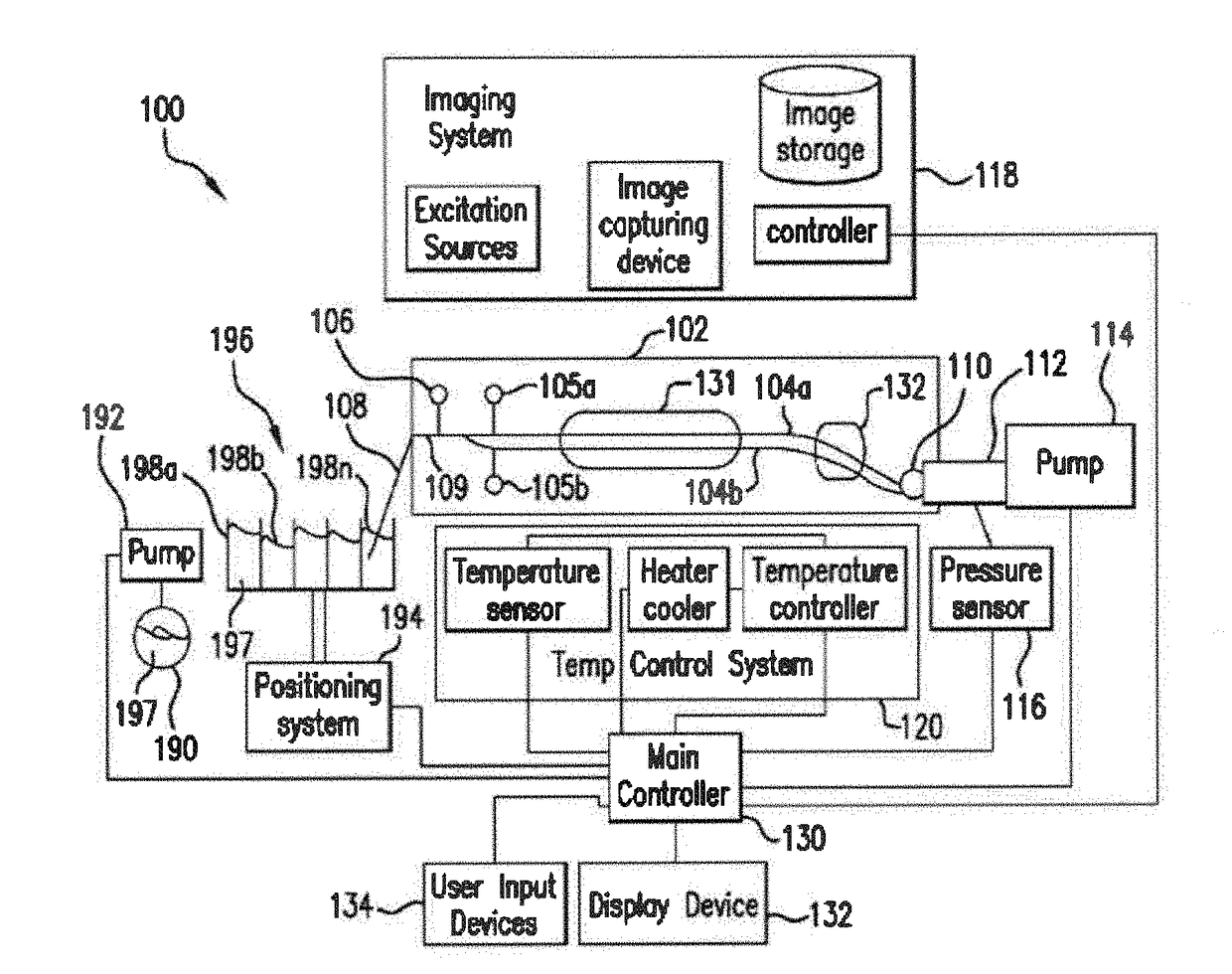
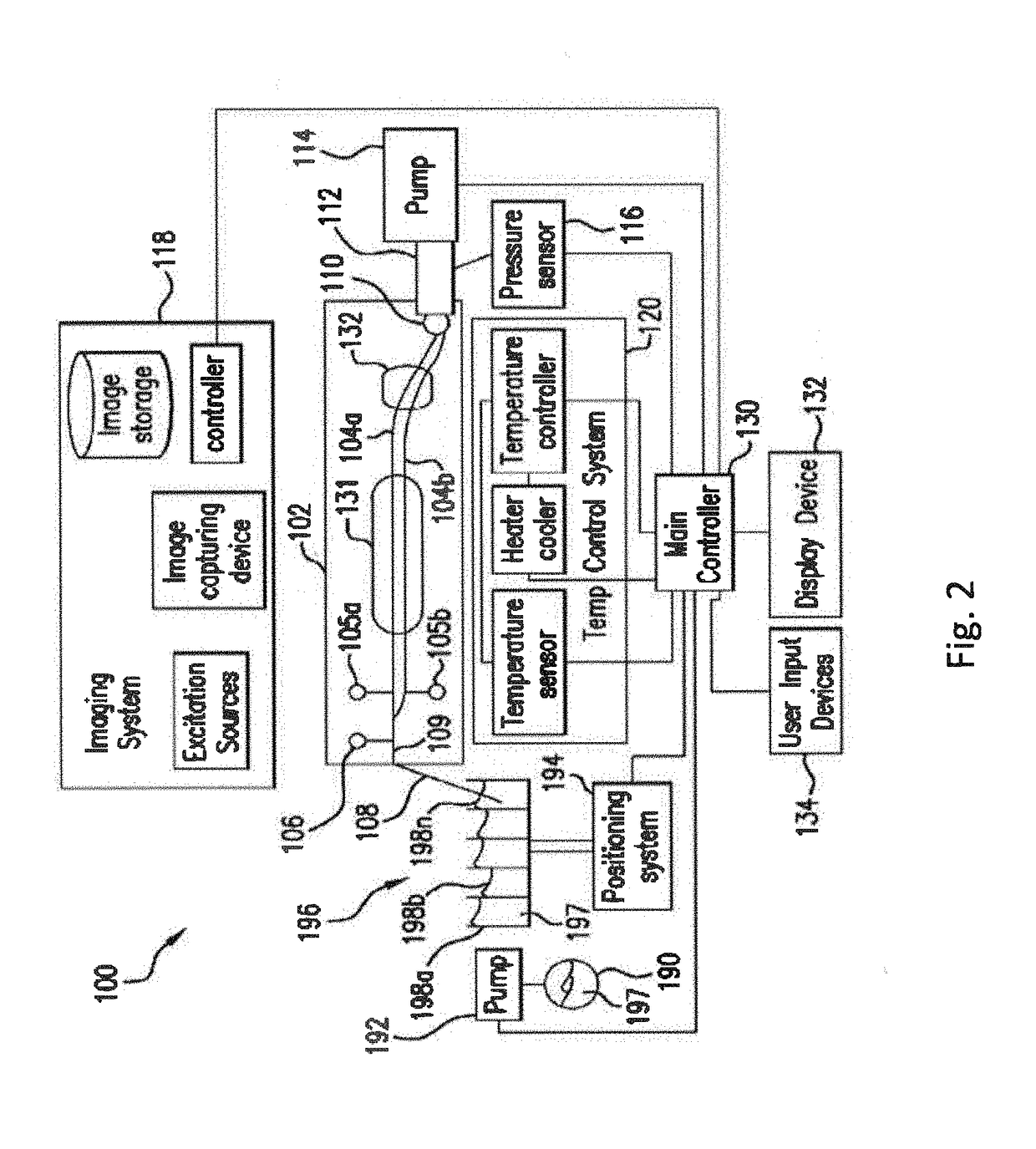
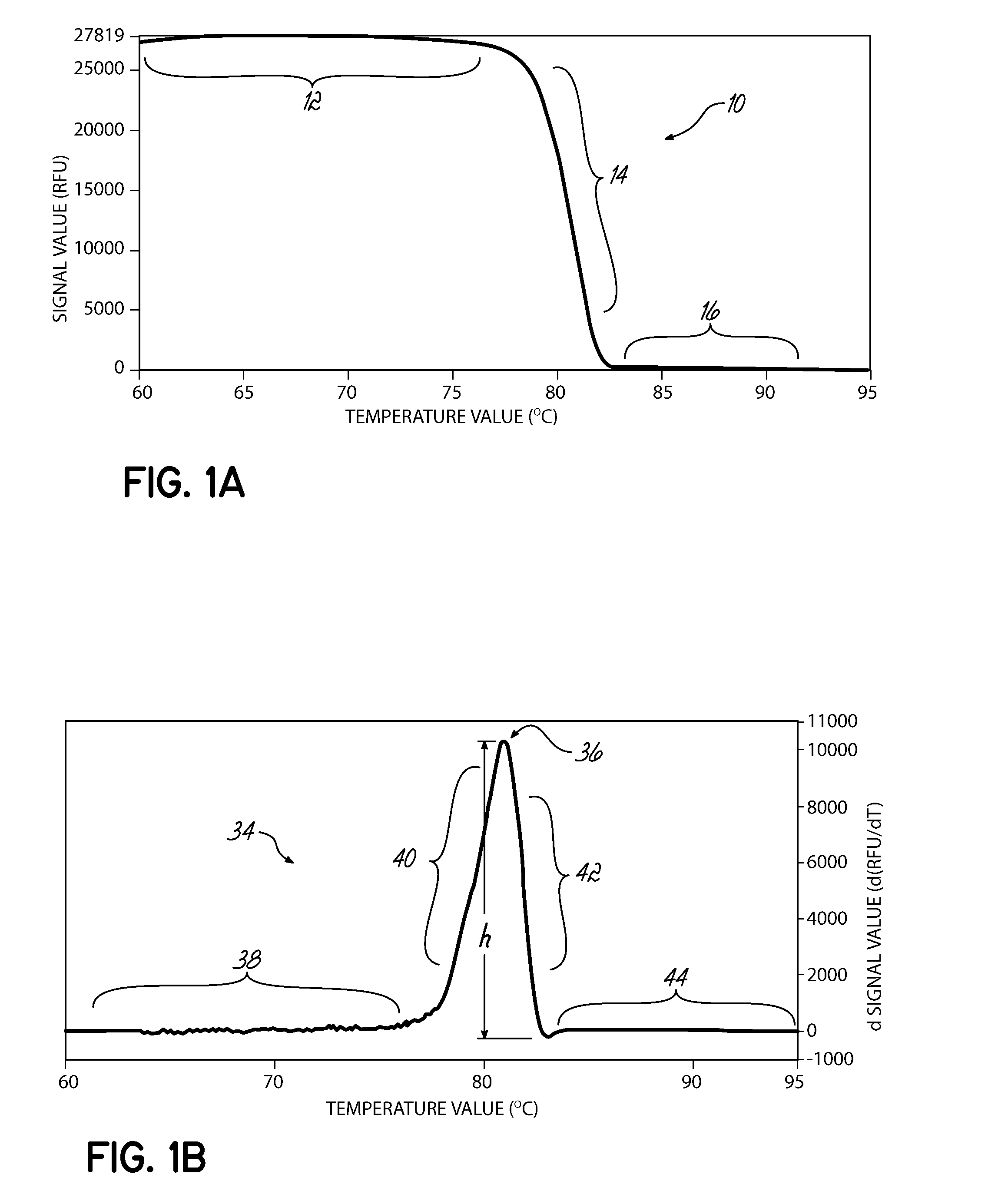
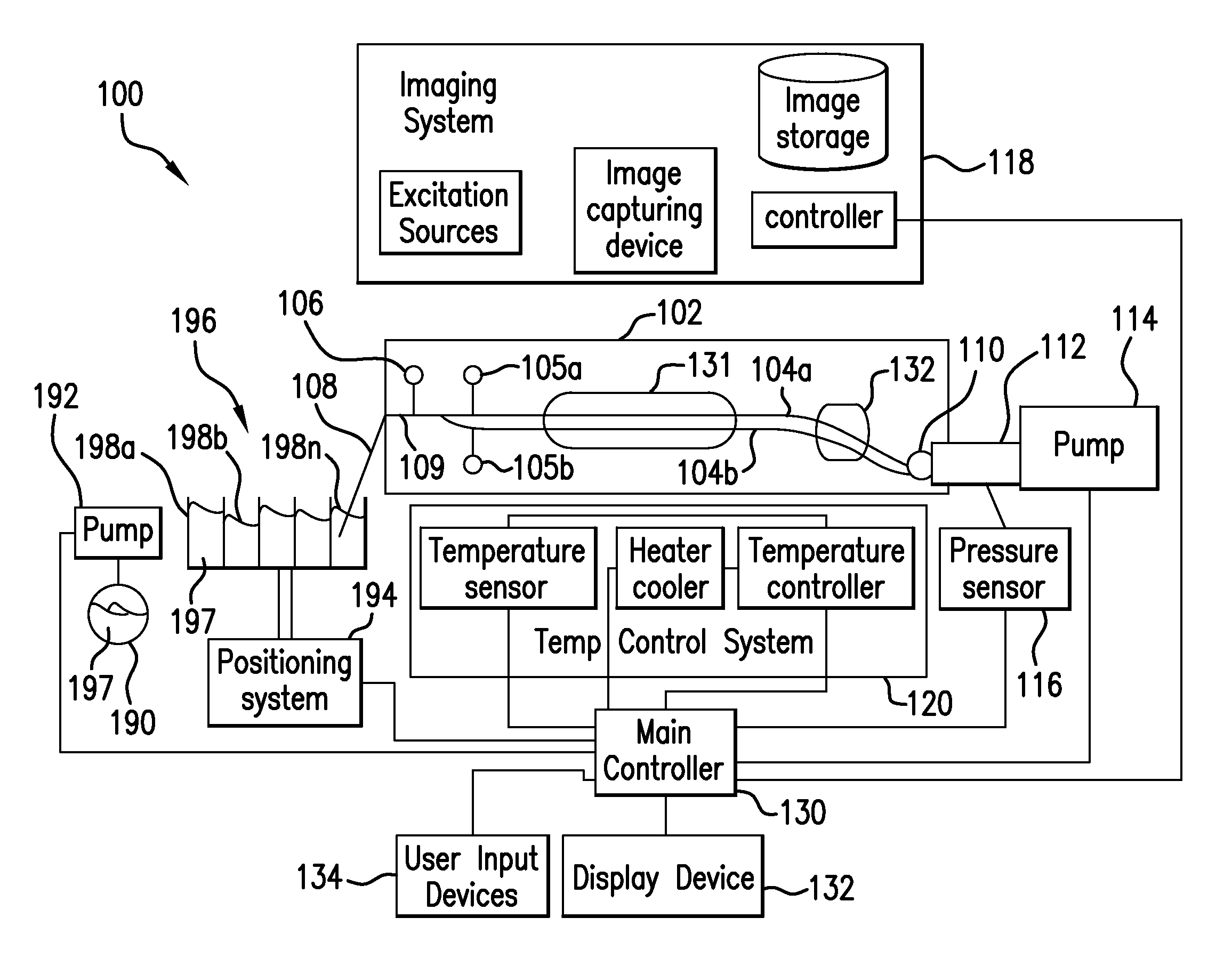
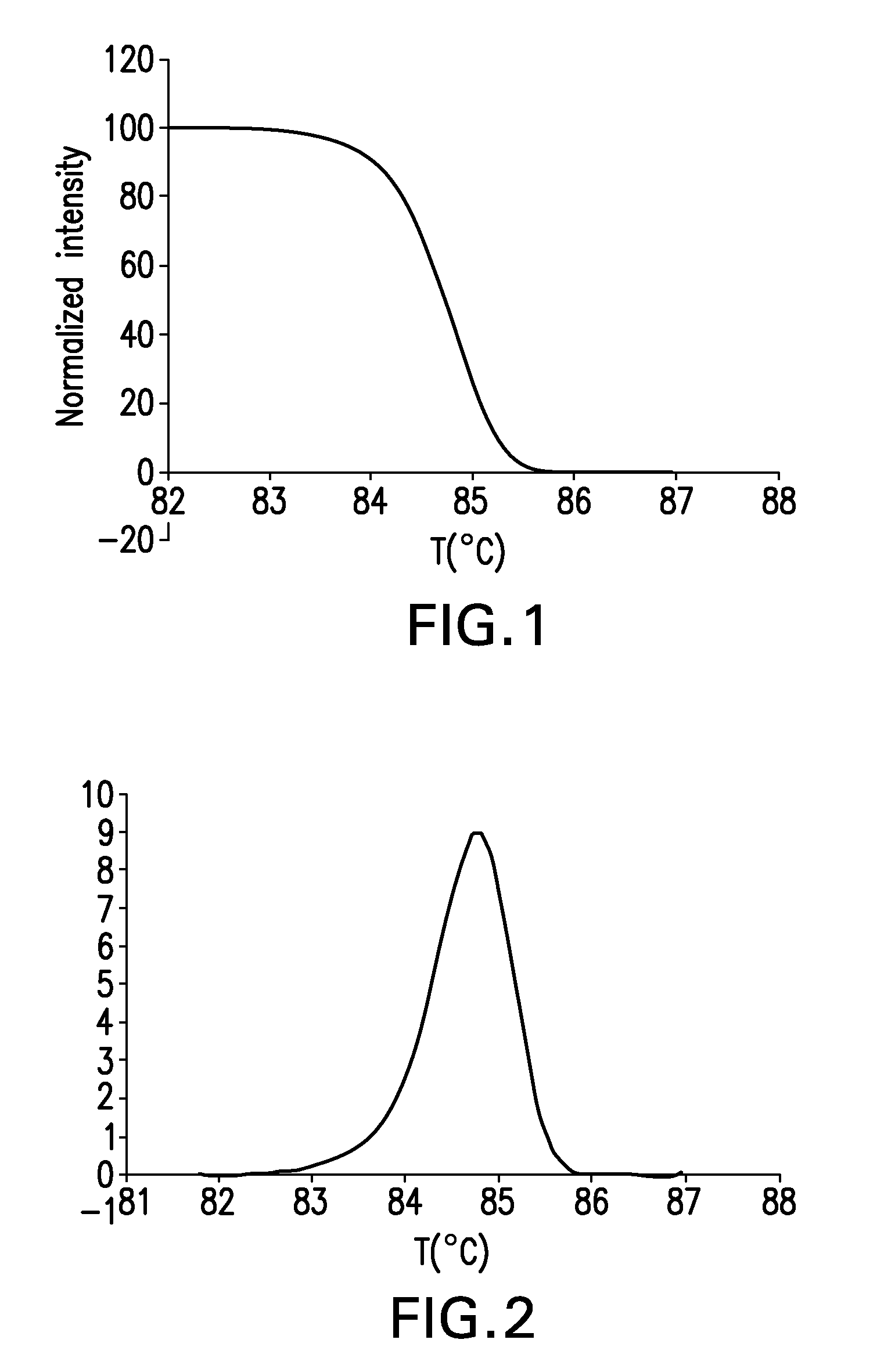
High-resolution melting analysis
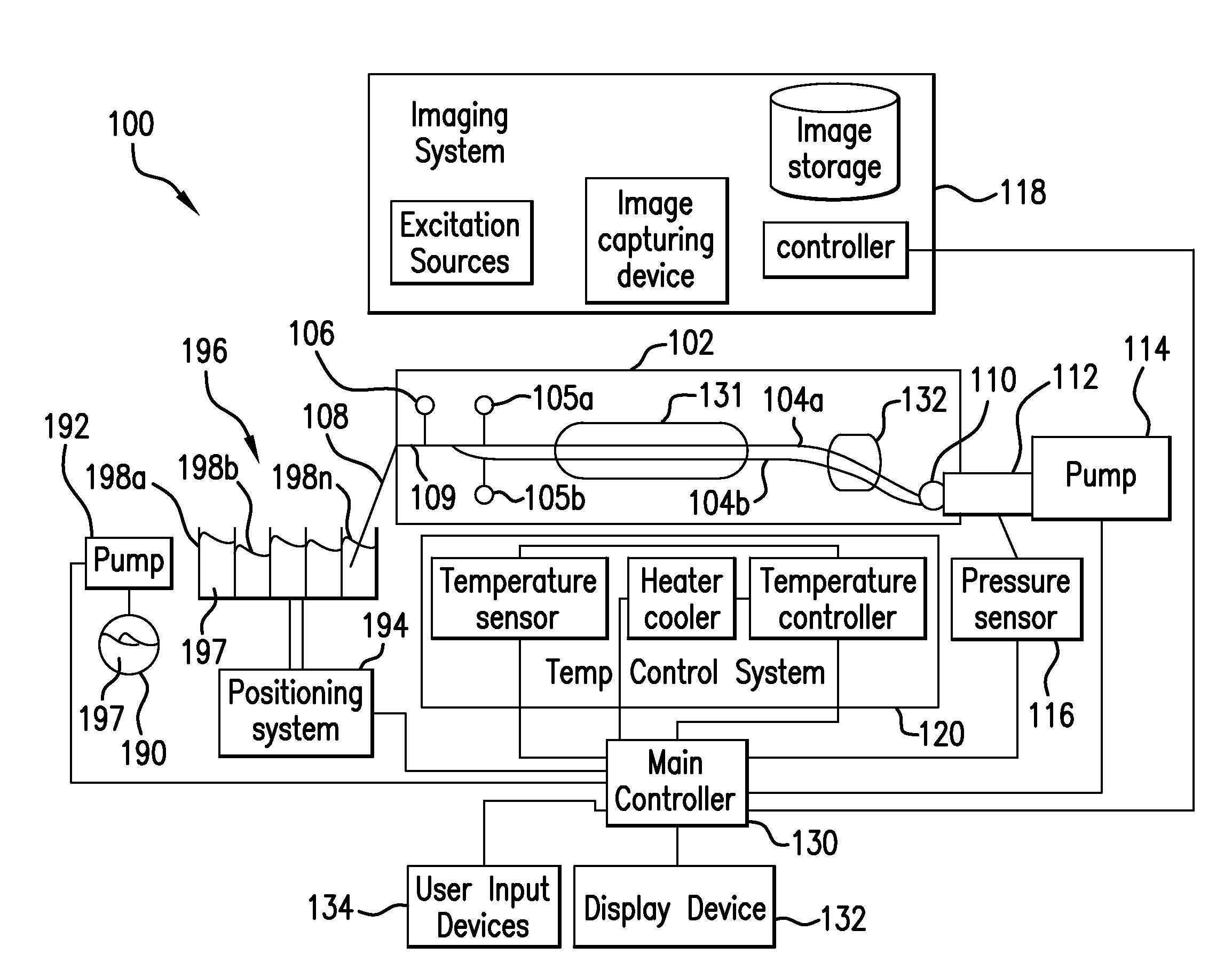
ActiveUS20090112484A1Heating or cooling apparatusMicrobiological testing/measurementNucleotidePhysical change
The present invention relates to methods and systems for the analysis of the dissociation behavior of nucleic acids and the identification of nucleic acids. In one aspect, methods and systems are disclosed for identifying a nucleic acid in a sample including an unknown nucleic acid and for detecting a single nucleotide polymorphism in a nucleic acid in a sample. In another aspect, methods and systems are disclosed for identification of a nucleic acid in a biological sample including at least one unknown nucleic acid by fitting denaturation data including measurements of a quantifiable physical change of the sample at a plurality of independent sample property points to a function to determine an intrinsic physical value and to obtain an estimated physical change function, and identifying the nucleic acid in the biological sample by comparing the intrinsic physical value for at least one unknown nucleic acid to an intrinsic physical value for a known nucleic acid.
Owner:CANON US LIFE SCIENCES INC
One step method inverse transcription PCR kit for detecting and differentiating Zika viruses and detection method thereof
InactiveCN105936946AImprove accuracyIncrease credibilityMicrobiological testing/measurementDNA/RNA fragmentationPathogenMelt temperature
The invention provides a primer, probe, and kit for detecting and differentiating Zika viruses through one step method inverse transcription PCR, a detection method and applications thereof. The PCR kit can be sensitively, specifically, efficiently, and rapidly detect and differentiate two genotypes of Zika viruses namely Asia type and Africa type at the same time. The principle of the kit is that on the basis of an inverse transcription PCR technology, a one step method FRET-PCR system, which can simultaneously detect and differentiate two Zika viruses, is established. The system is built on the basis of two genotypes of Zika viruses and whole gene sequences of other pathogens having a high homology with two genotypes of Zika viruses; a relatively conservative section is selected to design the primer and probe, through specific amplification, positive samples can be detected and screened from clinical samples sensitively and rapidly; then according to the difference of melting temperature (TM) values in a high resolution melting curve, the Asia type and Africa type of Zika viruses can be differentiated credibly; operation of the kit and the detection method is convenient and efficient, and the kit and method are suitable for detecting a large amount of samples.
Owner:YANGZHOU UNIV
High resolution melting curve-based multi-SNP identification method
ActiveCN104131092AOvercoming the technical defect of poor stability in detecting SNPImprove stabilityMicrobiological testing/measurementElectrophoresesHigh Resolution Melt Analysis
The invention belongs to the technical fields of the agriculture and the biology, and concretely discloses a high resolution melting curve-based multi-SNP identification method. The high resolution melting curve-based multi-SNP identification method is provided by combining multi-PCR, nested PCR and allele specific PCR against the limitations of present SNP identification. The utilization of the multi-PCR realizes 2-4 gene fragment amplification, improves the experiment efficiency, and reduces the experiment cost; the utilization of the nested PCR improves the specificity of amplified fragments, obtains a high quality and uniform concentration DNA template, and improves the stability of an HRM technology; the utilization of the allele specific PCR realizes SNP site discrimination, avoids fluorescent marker set or complicated electrophoresis and enzyme cutting operation; and the method comprehensively utilizing the multi-PCR, nested PCR and allele specific PCR realizes multi-SNP batch identification reduces the experiment cost.
Owner:SOUTH CHINA AGRI UNIV
Method for detecting vibrio parahemolyticus through combination of unlabelled fluorescent PCR (Polymerase Chain Reaction) and HRMA (High Resolution Melting Analysis)
ActiveCN103184279AGood repeatabilityEasy to detectMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceVibrio parahemolyticus
The invention discloses a method for detecting vibrio parahemolyticus through combination of unlabelled fluorescent PCR and HRMA, which is characterized by including the following steps: firstly, a primer pair is designed according to a TLH (thermolabile hemolysin) gene of the vibrio parahemolyticus; secondly, after DNA (Deoxyribonucleic acid) samples are extracted, unlabelled fluorescent PCR amplification is performed by utilizing the designed primer pair; and thirdly, an amplification curve of a PCR amplification product and the HRMA are analyzed by application software. The purposes of the method are to overcome defects of the prior art and provide a method, which is simple, convenient and fast to operate, accurate in detection result and low in usage cost, for detecting the vibrio parahemolyticus through the combination of the unlabelled fluorescent PCR and the HRMA.
Owner:邳州市博睿投资管理有限公司
Integrated purification and measurement of DNA methylation and co-measurement of mutations and/or mRNA expression levels in an automated reaction cartridge
PendingCN110267744AHeating or cooling apparatusMicrobiological testing/measurementNucleic acid sequencingSulfite
Methods of determining methylation of DNA are provided. In one illustrative, but non-limiting embodiment the method comprises i) contacting a biological sample comprising a nucleic acid to a first matrix material comprising a first column or filter where said matrix material binds and / or filters nucleic acids in said sample and thereby purifies the DNA; ii) eluting the bound DNA from the first matrix material and denaturing the DNA to produce eluted denatured DNA; iii) heating the eluted DNA in the presence of bi sulfite ions to produce a deaminated nucleic acid; iv) contacting said deaminated nucleic acid to a second matrix material comprising a second column to bind said deaminated nucleic acid to said second matrix material; v) desulphonating the bound deaminated nucleic acid and / or simultaneously eluting and desulphonating the nucleic acid by contacting the deaminated nucleic acid with an alkaline solution to produce a bi sulfite converted nucleic acid; vi) eluting said bi sulfite converted nucleic acid from said second matrix material; and vii) performing methylation specific PCR and / or nucleic acid sequencing, and / or high resolution melting analysis (HRM) on said bisulfite-converted nucleic acid to determine the methylation of said nucleic acid, wherein at least steps iv) through vi) are performed in a single reaction cartridge.
Owner:CEPHEID INC
High-resolution melting analysis
InactiveUS20090112481A1Heating or cooling apparatusMicrobiological testing/measurementHigh Resolution Melt AnalysisComputational biology
The present invention relates to methods and systems for the analysis of the dissociation behavior of nucleic acids and the identification of nucleic acids. In one aspect, methods and systems are disclosed for resolving a denaturation curve of a sample containing a first and second nucleic acid into a resolved denaturation curve for the first nucleic acid and a resolved denuration curve for the second nucleic acid.
Owner:CANON US LIFE SCIENCES INC
Zika virus detection kit based on high resolution melting analysis and detection method thereof
InactiveCN110229931ADetection sensitivityDetection characteristicsMicrobiological testing/measurementDNA/RNA fragmentationZika virusNucleic acid detection
The invention relates to the field of nucleic acid detection, and discloses a Zika virus detection kit based on high resolution melting analysis and a detection method thereof. The invention is characterized in that specific primers are designed according to Zika virus NS3 gene sequences, Zika virus nucleic acid is detected by using an optimized high resolution melting reaction system, so that theZika virus detection kit is simple in operations, fast in speed and free of the need of post-processing on PCR products and capable of detecting the difference of single base and truly realizing theclosed-tube operation.
Owner:山东国际旅行卫生保健中心
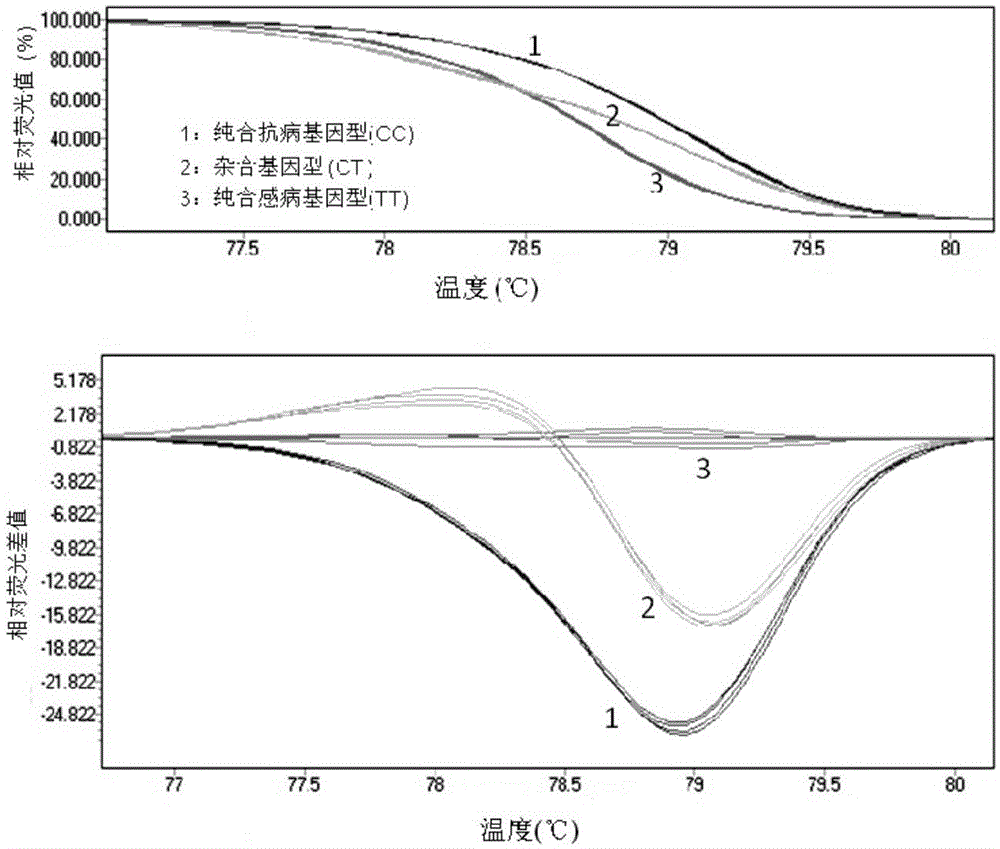
Transforming growth factor-beta (TGF-beta) type I receptor gene of chlamys farreri and single nucleotide polymorphism (SNP) locus of TGF-beta type I receptor gene
InactiveCN102899330AImprove seed selection efficiencyMicrobiological testing/measurementClimate change adaptationComplementary deoxyribonucleic acidMolecular genetics
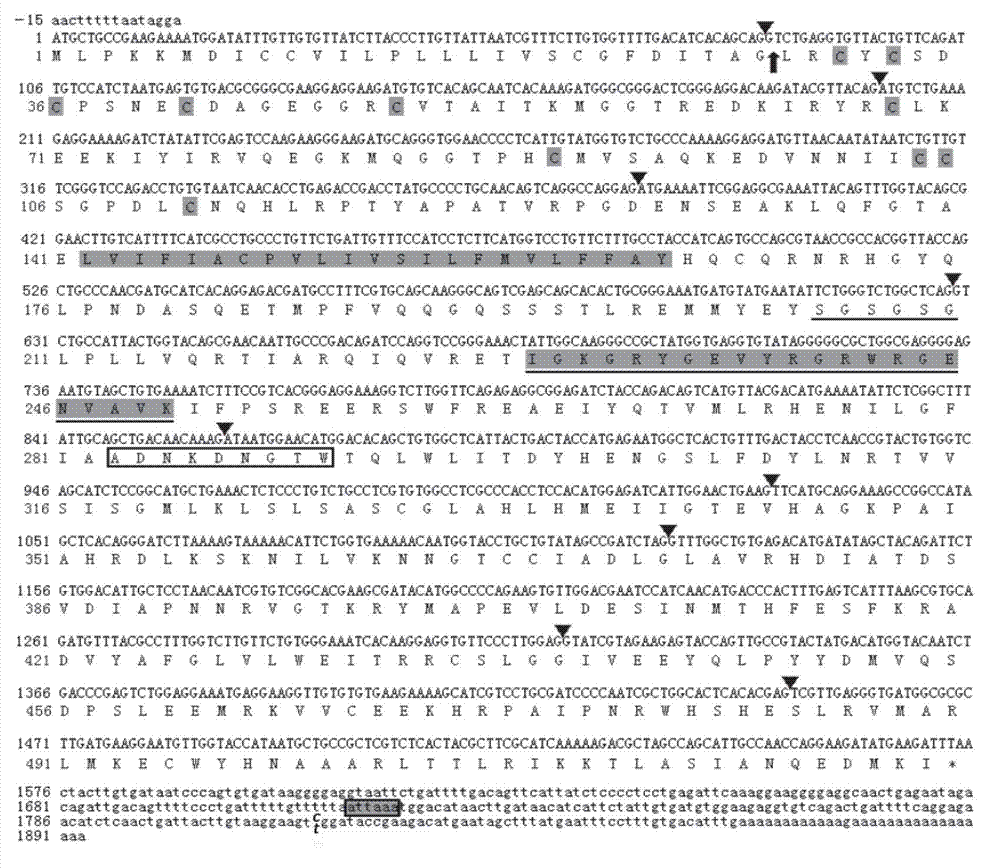
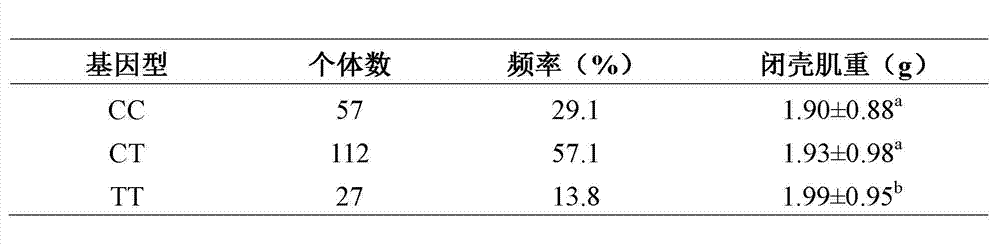
The invention relates to the cloning of a transforming growth factor-beta (TGF-beta) superfamily type I receptor gene Tgfbrl1 of chlamys farreri in a molecular genetic marker technology, a screening and typing technology of a single nucleotide polymorphism (SNP) locus which is relevant with the weight of adductor muscles in the gene and a method for the application of the gene to the breeding of the high-yield chlamys farreri. A total-length complementary deoxyribonucleic acid (cDNA) sequence of the Tgfbr1 gene of the chlamys farreri is obtained by utilizing a homology-based cloning strategy; an SNP lotus is discovered by blastn comparison, and primers and a probe are designed for the locus; and SNP typing is performed in a natural population of the chlamys farreri by using a high-resolution melting curve technology, and the weight of the adductor muscle of an individual is measured. Statistic analysis displays that the loci c.1815C>T are obviously relevant with the weight of the adductor muscles of the chlamys farreri, and the weight of the adductor muscles of individuals with TT genetypes is obviously higher than that of the adductor muscles of individuals with CC and CT genetypes. In the selective breeding process of the high-yield chlamys farreri, the breeding candidate colonies of the chlamys farreri can be subjected to c.1815C>T typing, and the individuals of which the loci c.1815C>T are TT genetypes are used as a breeding parent preferably by combining typing information of other loci which are relevant with growth properties.
Owner:OCEAN UNIV OF CHINA
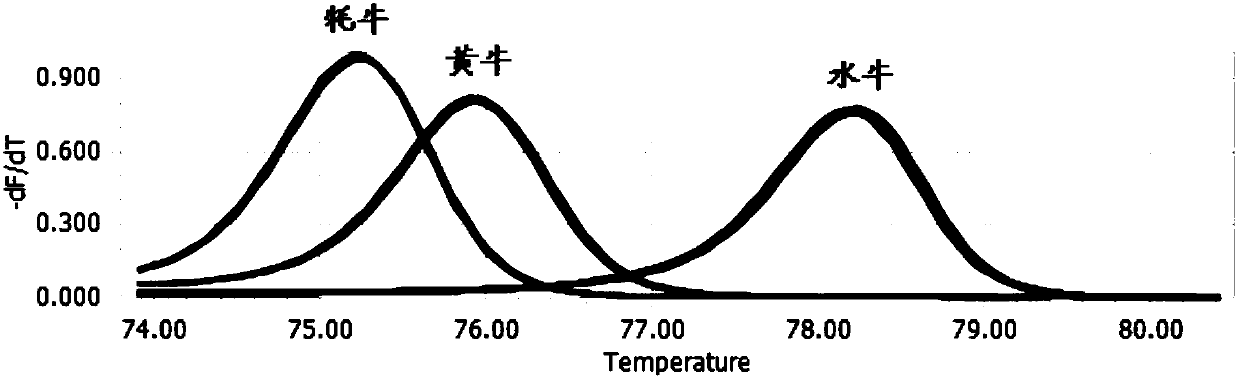
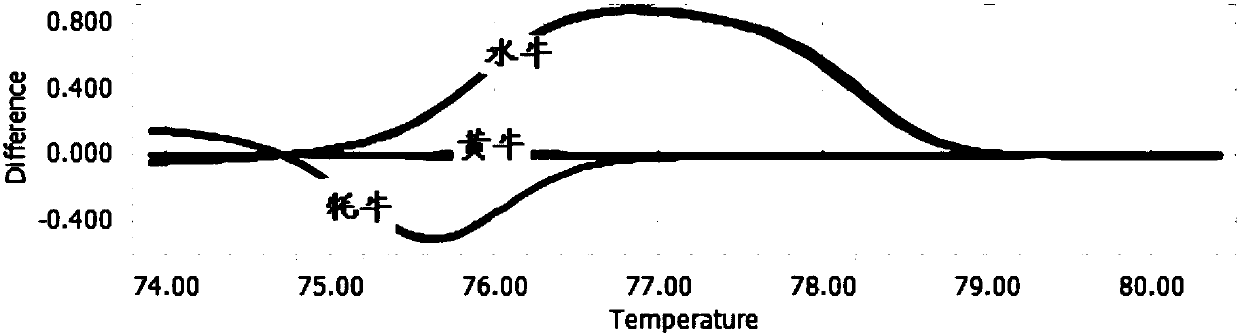
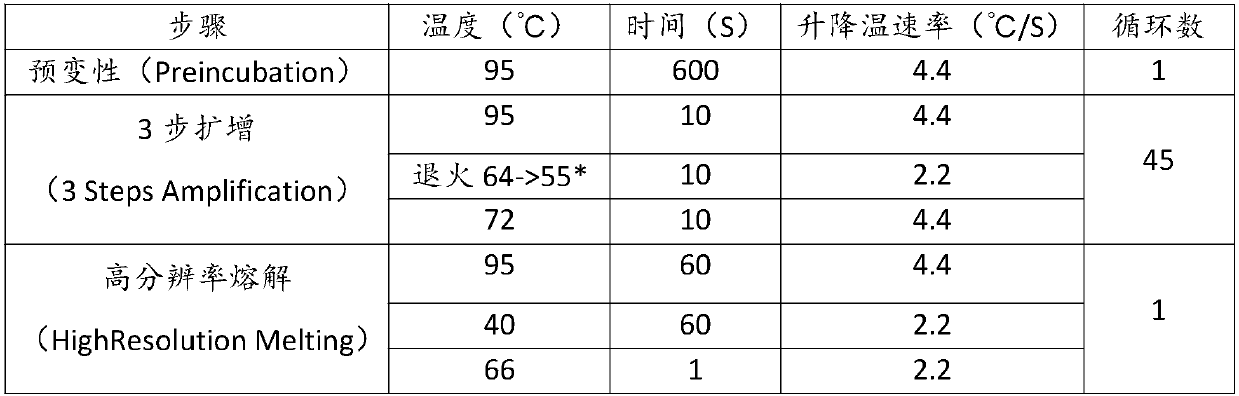
Detection primers and method for yak, cattle and buffalo derived components in beef food
PendingCN107858442AReduce consumable costsShorten detection timeMicrobiological testing/measurementDNA/RNA fragmentationWater buffaloGenotyping Techniques
The invention discloses detection primers and a method for yak, cattle and buffalo derived components in beef food. The primers comprise a primer GTB-F and a primer GTB-R, wherein the nucleotide sequence of the GTB-F is shown as SEQ ID NO.1; the nucleotide sequence of the GTB-R is shown as SEQ ID NO.2. The method comprises the following steps: carrying out quantitative PCR (Polymerase Chain Reaction) amplification on a to-be-detected sample by using the primers to obtain QPCR amplification data; carrying out HRM (High Resolution Melting) data analysis and detection on an amplification productby utilizing quantitative PCR instrument bundled software. According to the detection primers and the method disclosed by the invention, a HRM curve genotyping technology is applied to detection of three different bovine derived materials, so that reagent consumption cost of relevant detection is greatly reduced; meanwhile, detection time is shortened; detection accuracy can be improved, and a false positive result is avoided.
Owner:SICHUAN LIGHT INDUSTRY RESEARCH AND DESIGN INST
High-resolution melting analysis
The present invention relates to methods and systems for the analysis of the dissociation behavior of nucleic acids and the identification of nucleic acids. In one aspect, methods and systems are disclosed for identifying a nucleic acid in a sample including an unknown nucleic acid and for detecting a single nucleotide polymorphism in a nucleic acid in a sample. In another aspect, methods and systems are disclosed for identification of a nucleic acid in a biological sample including at least one unknown nucleic acid by fitting denaturation data including measurements of a quantifiable physical change of the sample at a plurality of independent sample property points to a function to determine an intrinsic physical value and to obtain an estimated physical change function, and identifying the nucleic acid in the biological sample by comparing the intrinsic physical value for at least one unknown nucleic acid to an intrinsic physical value for a known nucleic acid.
Owner:CANON US LIFE SCIENCES INC
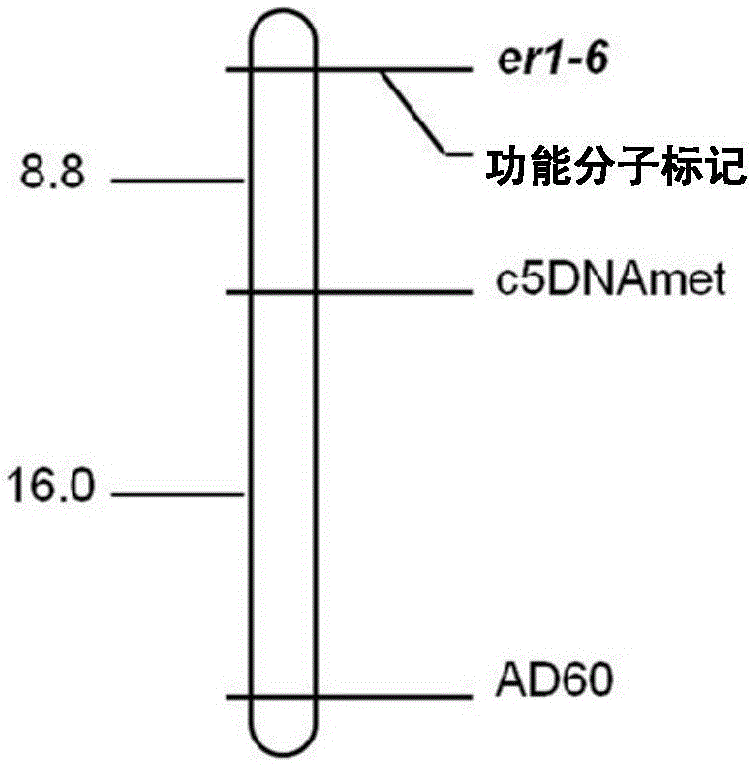
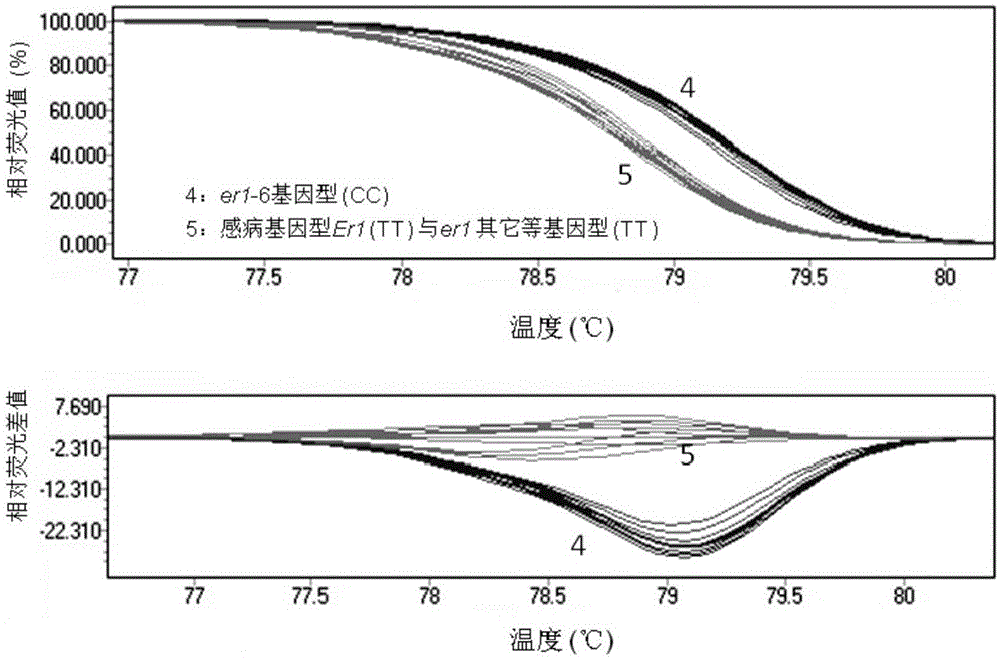
Molecular marker subjected to co-segregation with pea powdery mildew resistance allele er1-6 and application thereof
InactiveCN105039336AImprove throughputSimple and fast operationMicrobiological testing/measurementDNA/RNA fragmentationCandidate Gene Association StudyBiology
The invention provides a molecular marker subjected to co-segregation with a pea powdery mildew resistance allele er1-6. The molecular marker is located on a VI linkage group of a pea genetic map, and the genetic distance between the molecular marker and the allele er1-6 is 0cM. According to the single base difference (at the position of 1121, T-C) between a disease-resistant variety G0001778 containing the resistance allele er1-6 and an er1 candidate gene PsMLO1cDNA sequence of infected variety pea number 6, primers are designed on the two sides of an SNP mutation site, a high resolution melting curve analysis technology is used for developing the molecular marker subjected to co-segregation with the pea powdery mildew resistance allele er1-6, the marker is subjected to group detection of F2 and F3 which are derived by the disease-resistant variety G0001778 and the infected variety pea number 6 in a hybridization mode, and it is verified that the marker is a co-dominance functional marker subjected to co-segregation with the gene er1-6. Afterwards, the effectiveness is verified in pea powdery mildew resistance resource identification through the marker, the functional marker can be used for molecular marker-assisted selection breeding of the pea powdery mildew resistance, and therefore the breeding process is accelerated.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
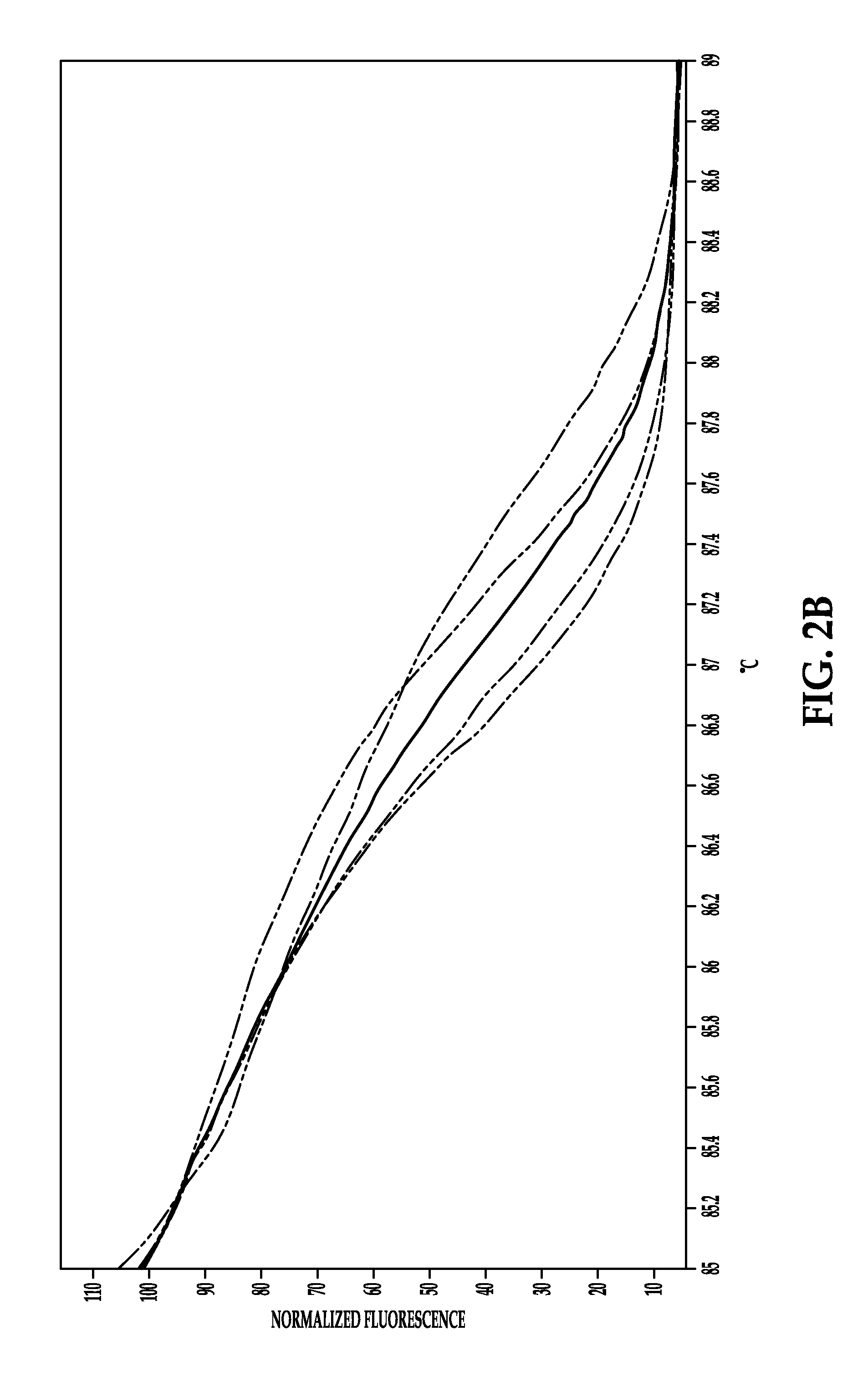
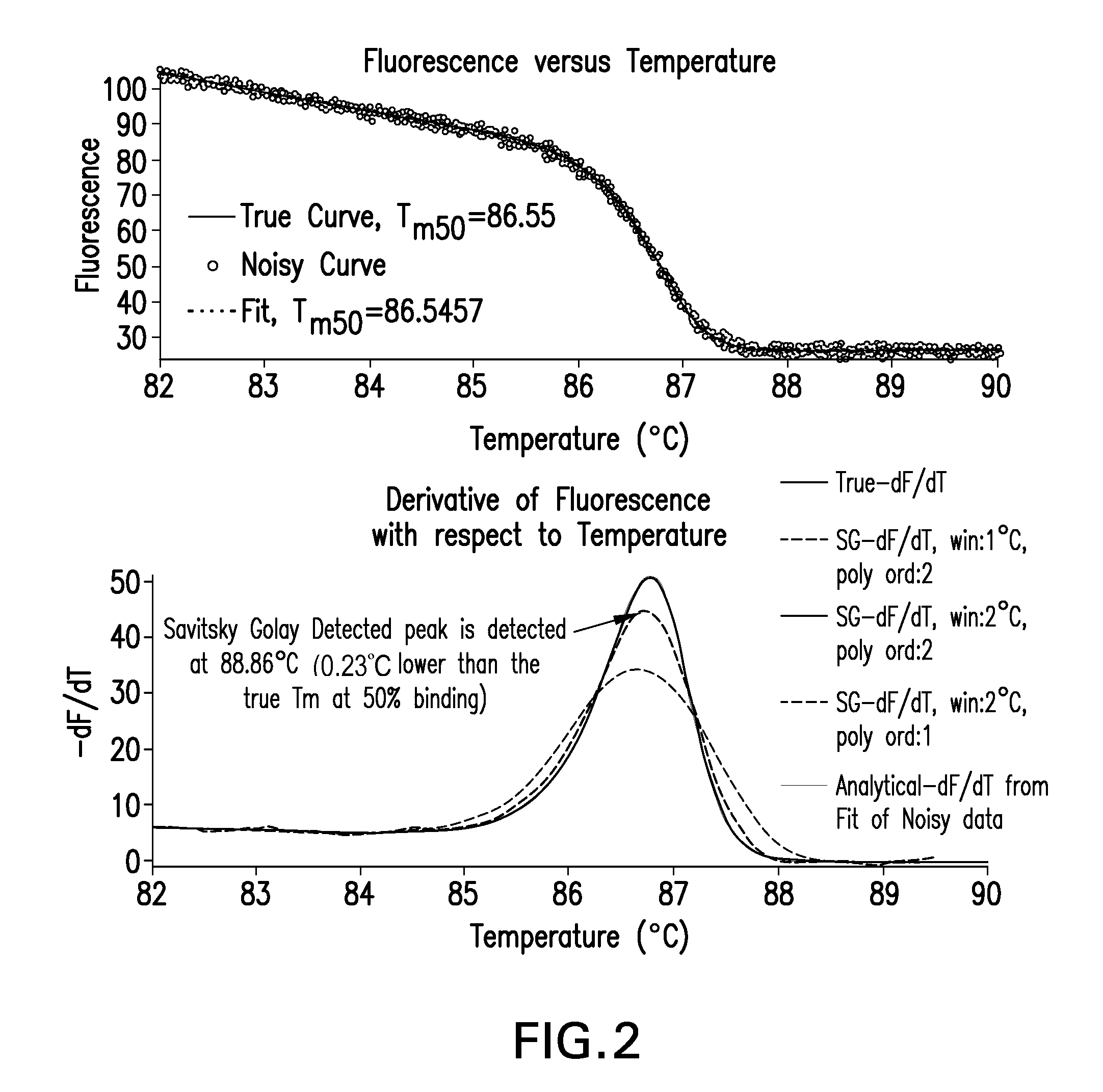
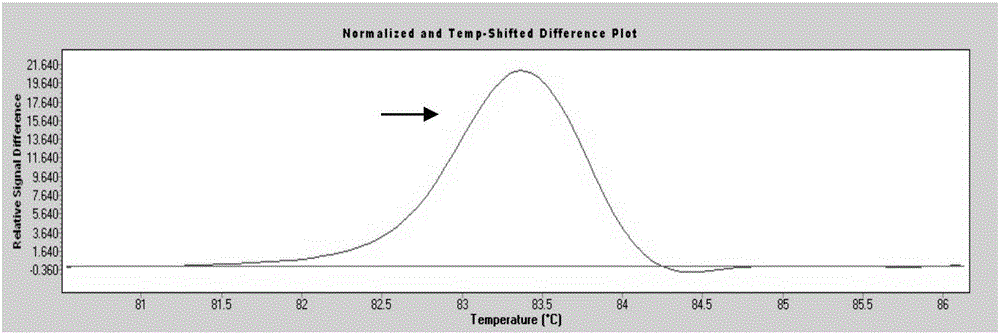
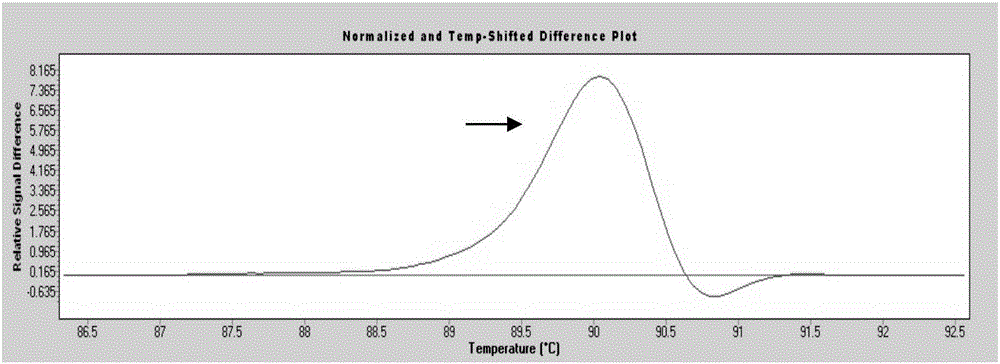
System and method for melting curve normalization
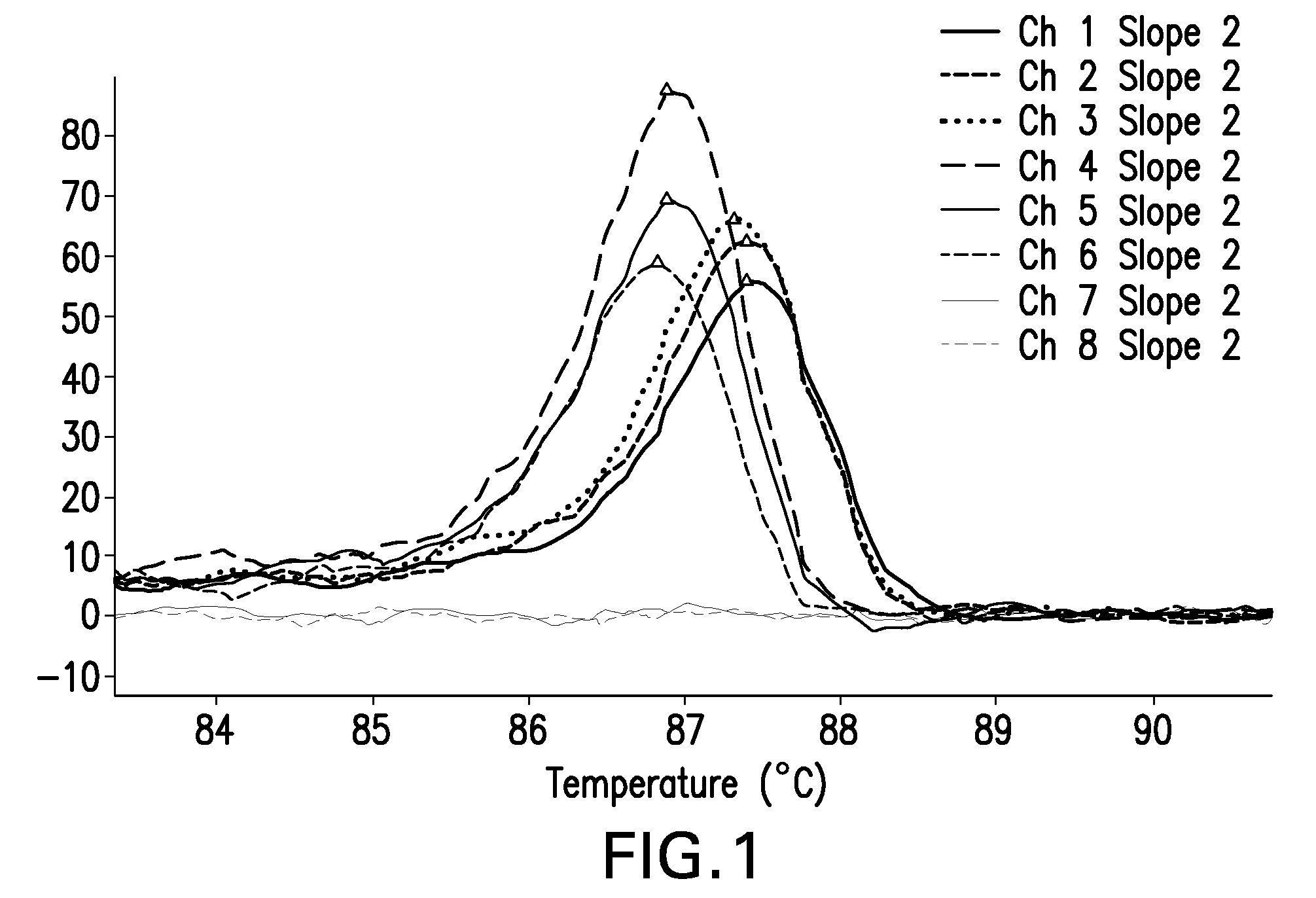
ActiveUS20170372002A1Increase temperatureMicrobiological testing/measurementData visualisationDNAHigh Resolution Melt Analysis
The present invention relates to methods for the analysis of nucleic acids present in biological samples, and more specifically to normalize a high resolution melt curve to assist in the identification of one or more properties of the nucleic acids. The present invention provides methods and systems that incorporate a background identification algorithm according to invention principles using raw melt curve data to identify reactions that are unrelated actual DNA melt reactions. Furthermore, a web-based application for analyzing experimental data is provided. The raw experimental data obtained from a variety of instruments is processed and analyzed on a server and presented to a user through a user interface (UI).
Owner:CANON US LIFE SCIENCES INC
Detection primer, amplification system and detection kit of microsatellite instability site-BAT 26 site
ActiveCN107475253ALow costSimplify operating proceduresMicrobiological testing/measurementDNA/RNA fragmentationMicrochiropteraTrue positive rate
The invention belongs to the field of biological medicine, and relates to a detection primer, amplification system and detection kit of a microsatellite instability site-BAT 26 site. The sequence of the primer is shown as SEQ ID NO:1 and SEQ ID NO:2. The primer is adopted to perform high-resolution melting curve method and HRM detection, and is high in sensitivity and specificity.
Owner:THE SIXTH AFFILIATED HOSPITAL OF SUN YAT SEN UNIV
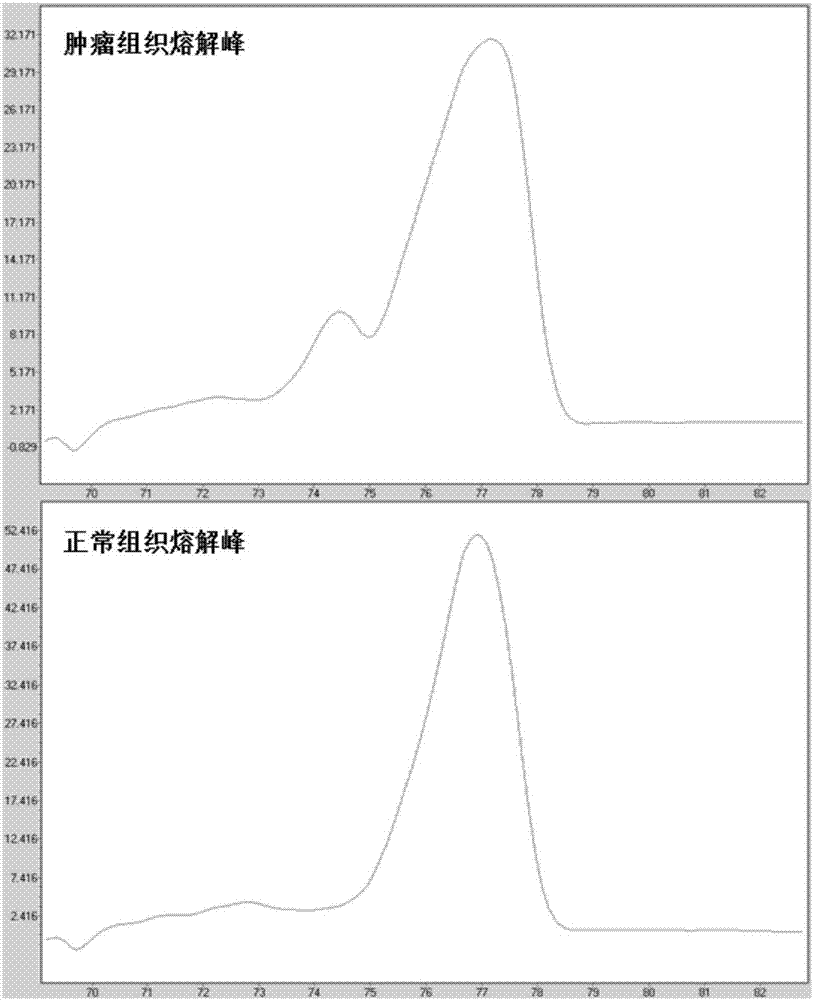
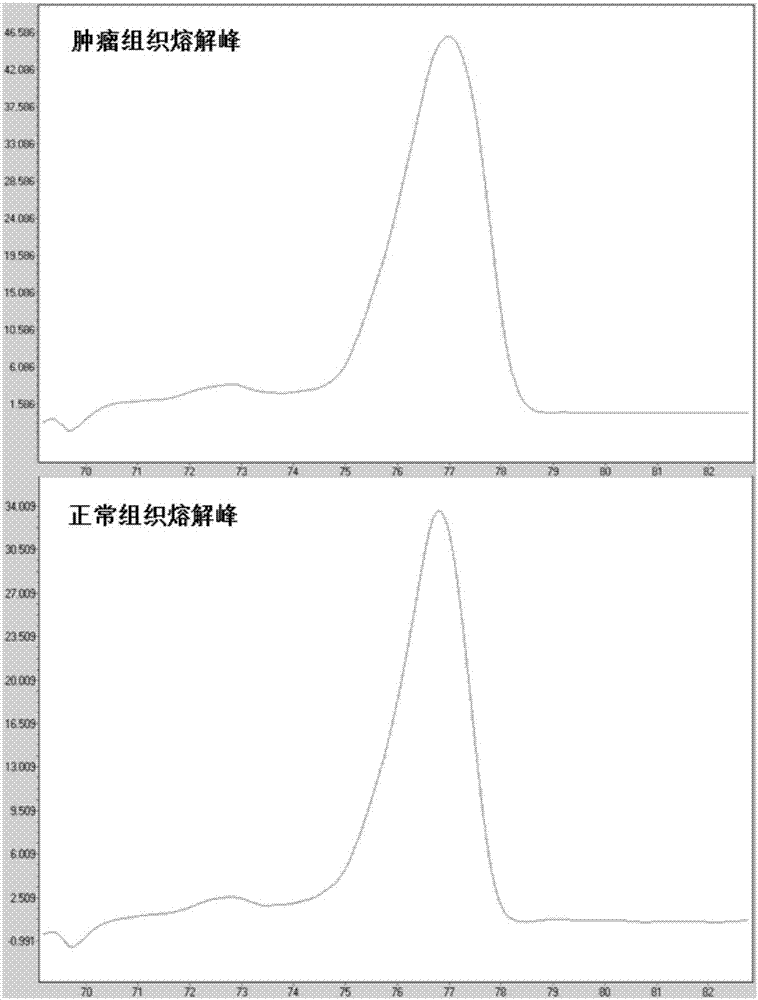
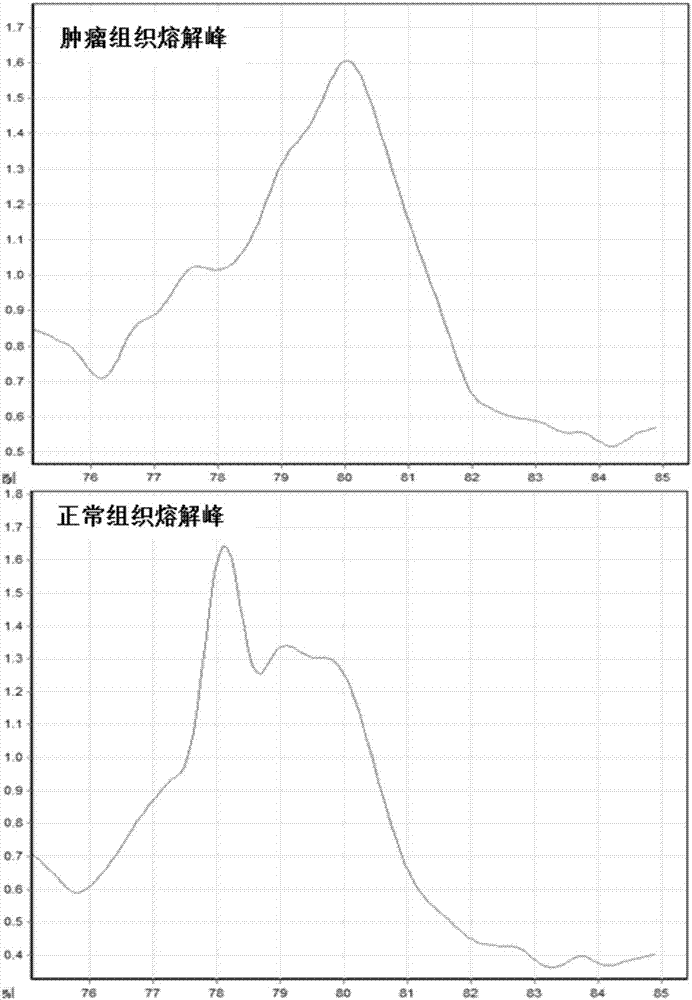
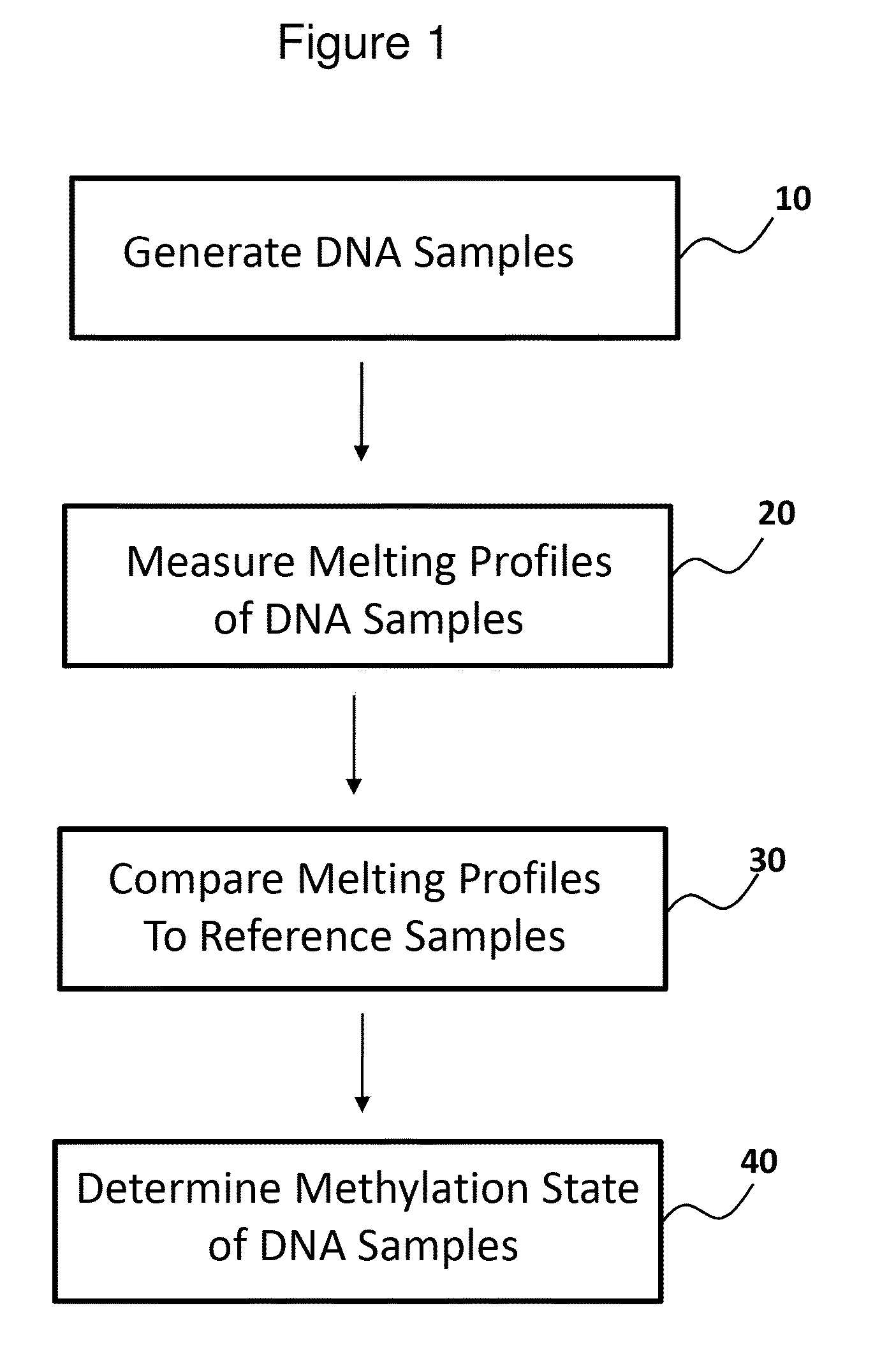
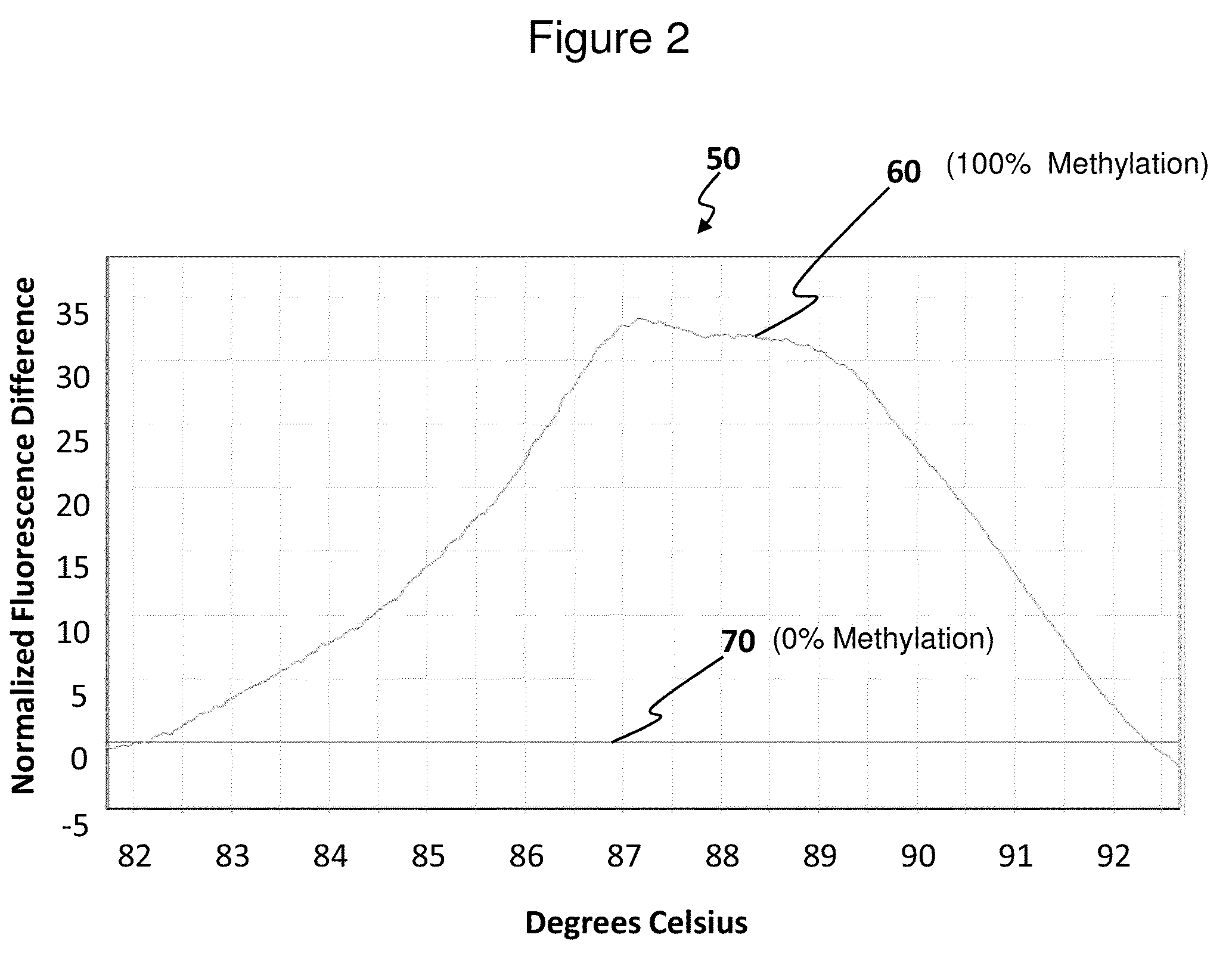
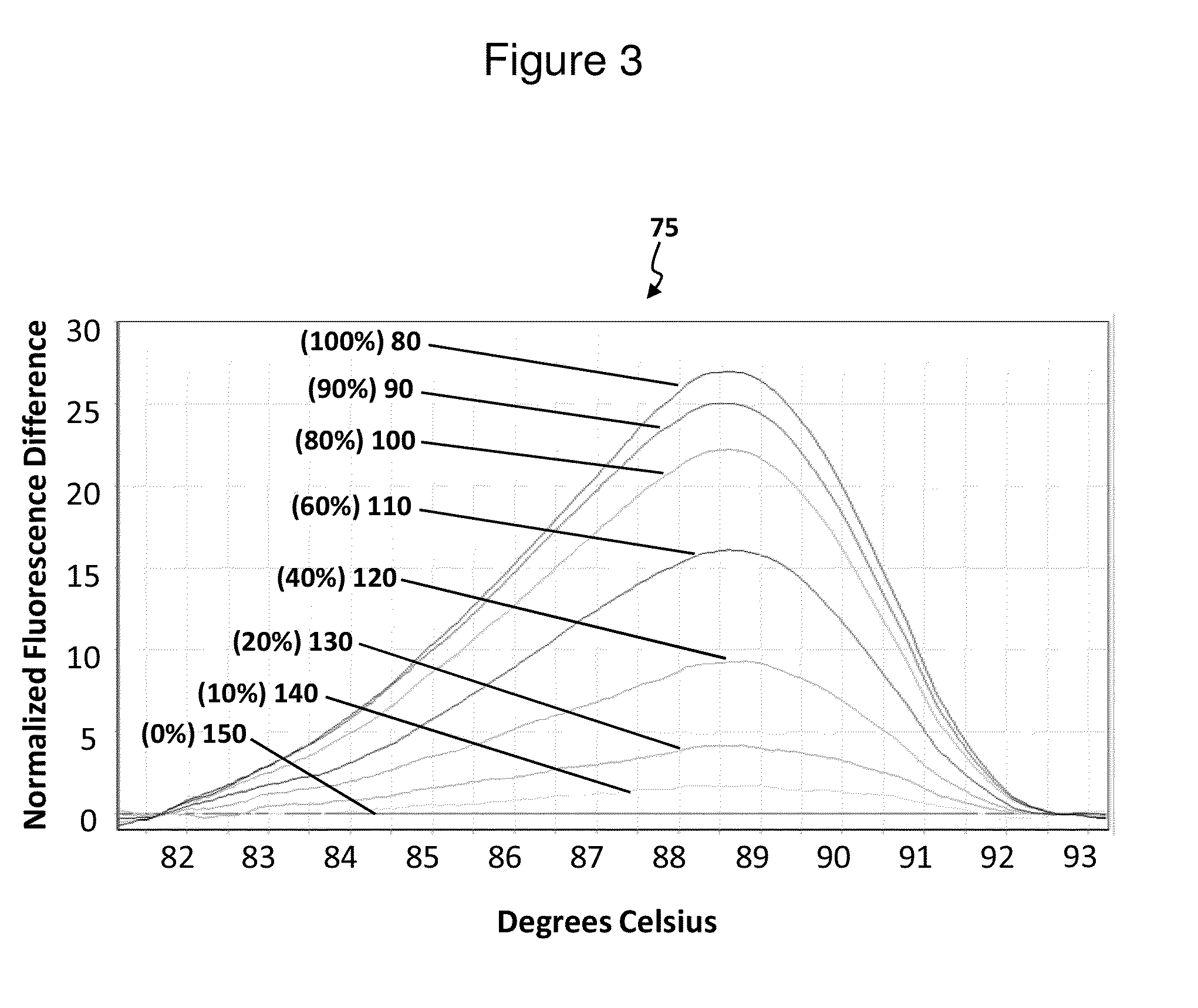
Methods for analysis of DNA methylation percentage
Methods are disclosed for determining the methylation state of DNA samples using melt analysis including high resolution melt analysis. Methods are also provided for determining methyltransferase activity using melt analysis including high resolution melt analysis. Additionally, kits of parts are provided.
Owner:ZYMO RES CORP
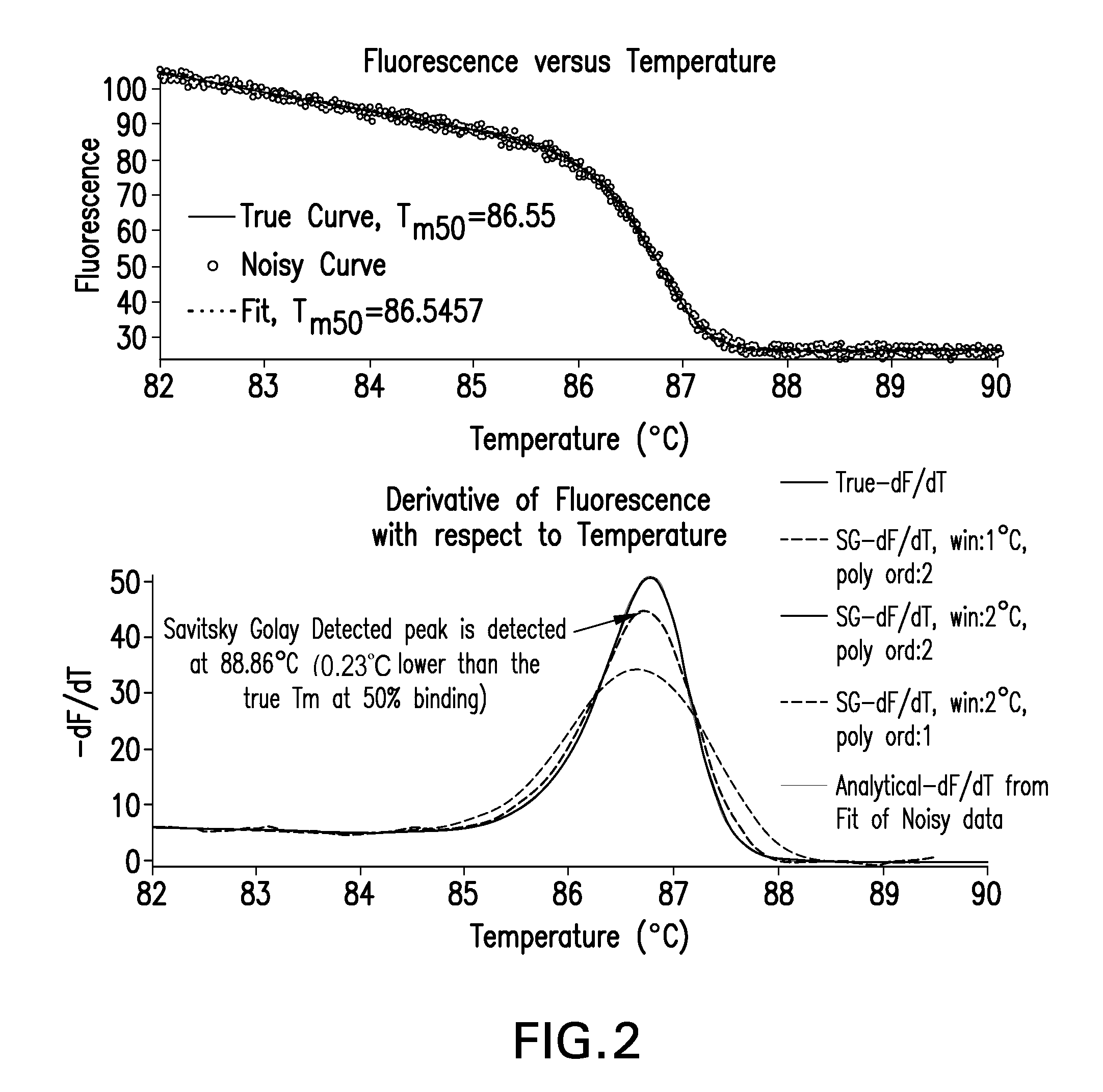
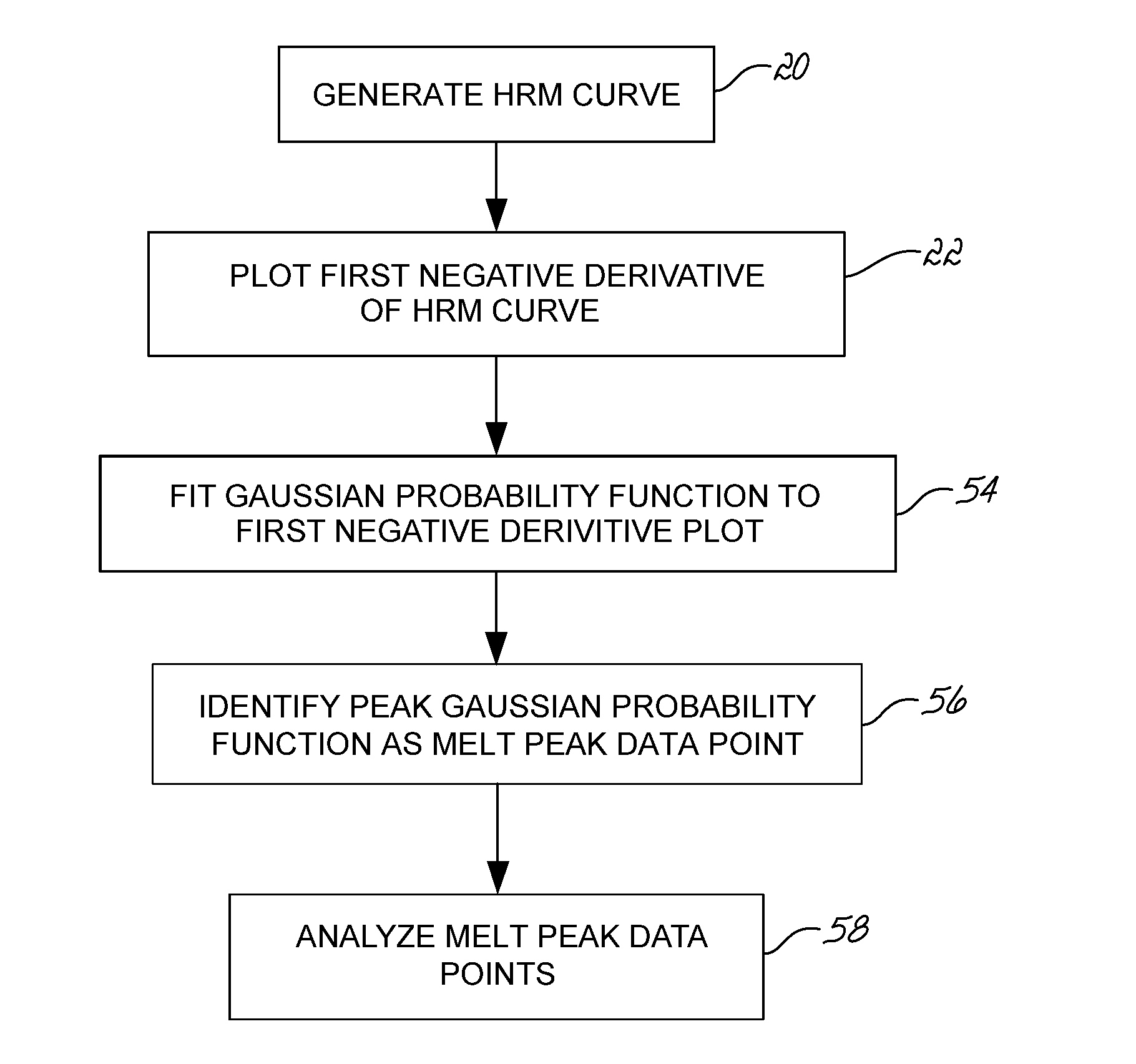
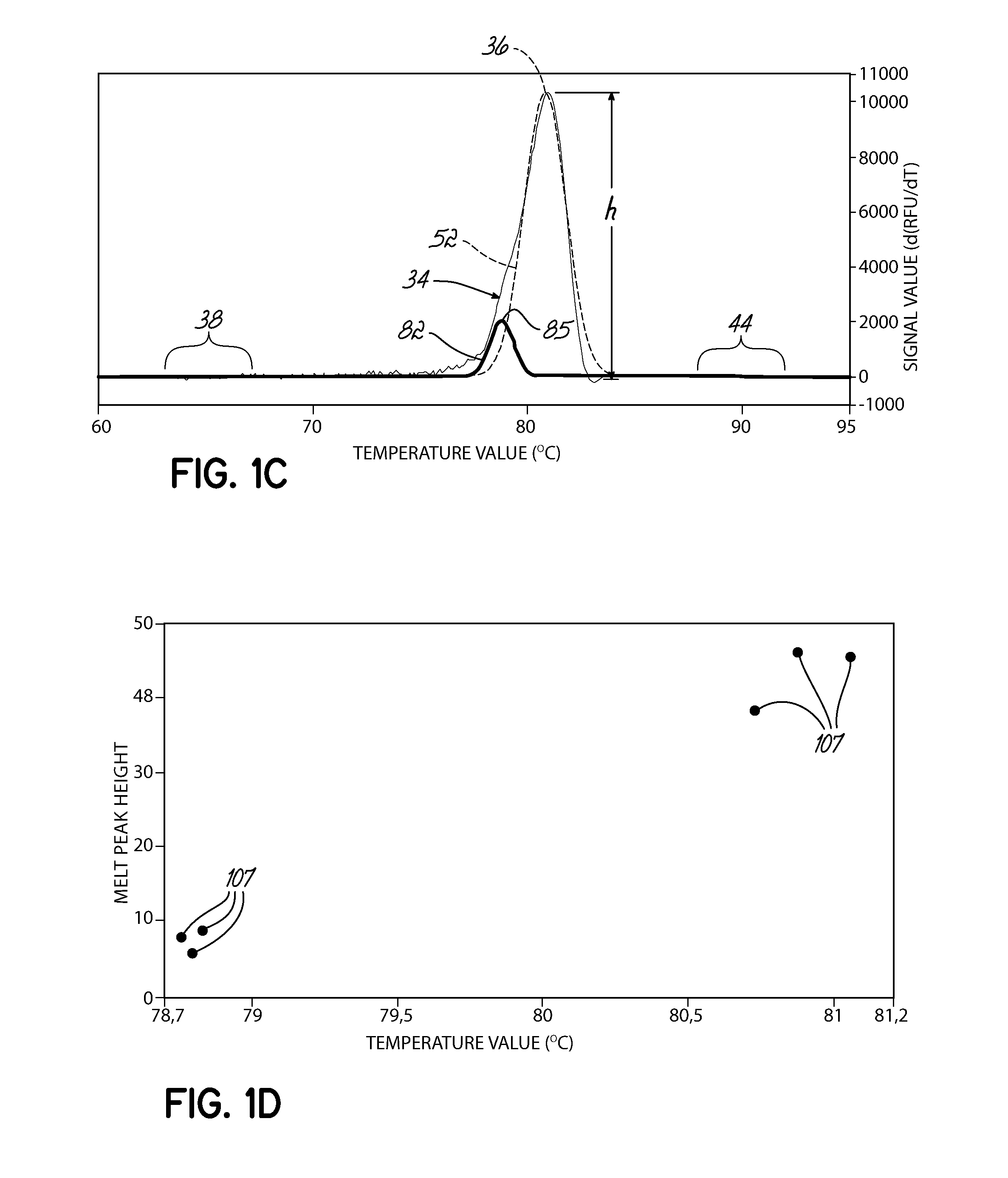
Methods and systems for high resolution melt analysis of a nucleic acid sequence
InactiveUS20130338928A1DiscriminationImprove discriminationMicrobiological testing/measurementData visualisationData setNucleic acid sequencing
Described herein are methods and systems for analyzing and visualizing HRM data from a double-stranded nucleic acid. The HRM data is generally characterized by a plurality of data points each including a signal value associated with the concentration of a double-stranded nucleic acid in a sample and a temperature value associated with a the temperature of the sample. Embodiments of the invention analyze the HRM curves from samples using the first negative derivative of the HRM curve or a virtual standard. The first negative derivative plot method may be used to identify the melting temperature of a homogenous double-stranded nucleic acid in a sample, as well as the presence and melting temperature of heterogeneous double-stranded nucleic acids in the sample. Data points associated with the melting temperature are plotted on a scatter plot for analysis. The virtual standard allows for visualization of HRM data across data sets.
Owner:THERMO FISCHER SCI OY
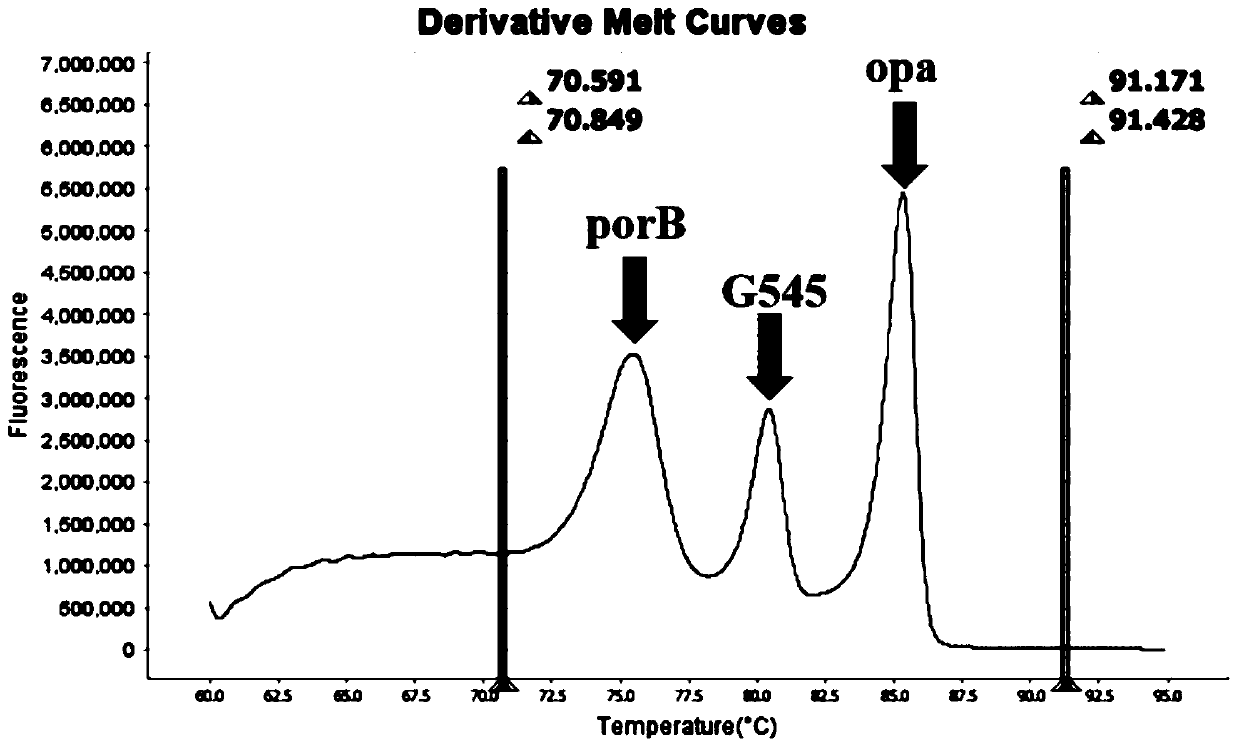
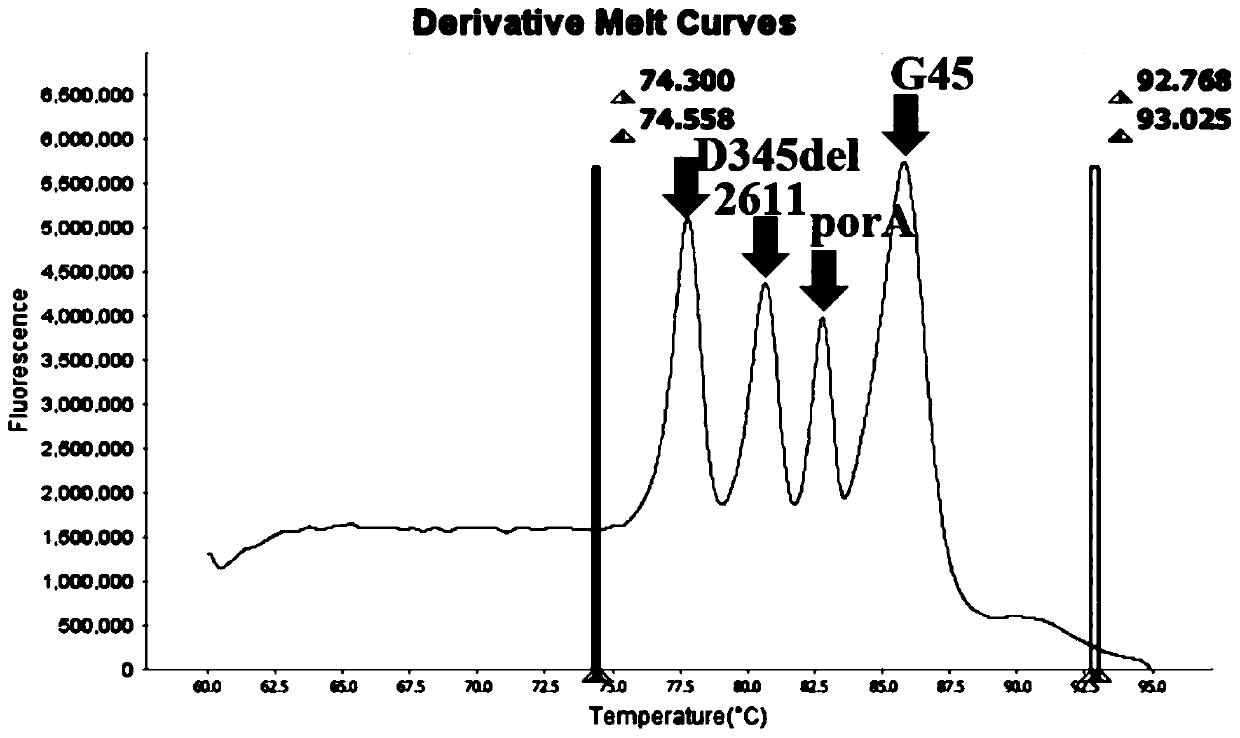
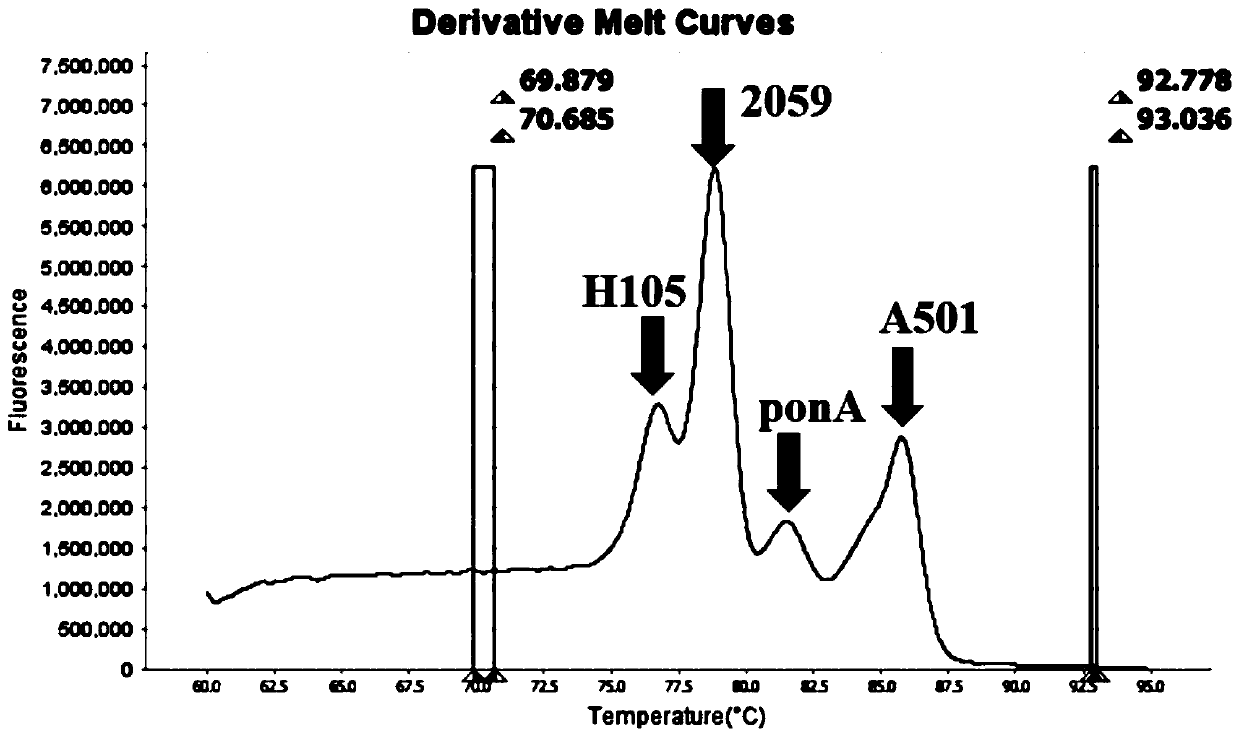
Method and kit for multiple detection of drug resistance sites of neisseria gonorrhoeae
PendingCN110643722AFast and Sensitive AnalysisImprove accuracyMicrobiological testing/measurementMicroorganism based processesHigh Resolution Melt AnalysisRelated gene
The invention belongs to the technical field of molecular biology detection, and relates to a detection method for drug resistance sites, in particular to a method and a kit for multiple detection ofthe drug resistance sites of neisseria gonorrhoeae. A provided specific primer for the multiple detection of the drug resistance sites of the neisseria gonorrhoeae selects drug-resistance-related genes of two drug combinations (ceftriaxone and azithromycin) as target genes for detection, and includes penA, ponA, porB, mtrR, and 23S rRNA. Based on high resolution melting curve analysis technique, the DNA demulsification process is monitored in real time through high resolution melting of PCR products, the mutation condition of the genes is analyzed according to the characteristic change of a melting curve, and thus a basis is provided for determining the drug-resistant condition of the neisseria gonorrhoeae.
Owner:USTAR BIOTECHNOLOGIES (HANGZHOU) CO LTD
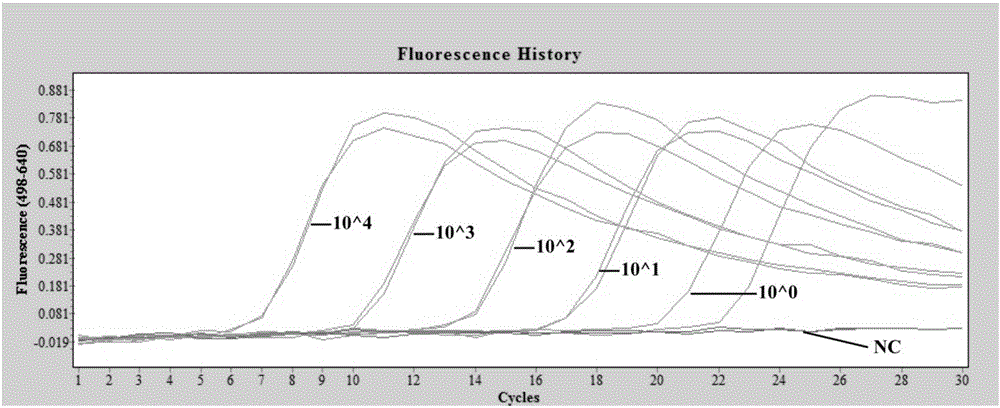
Digital PCR detection method
ActiveCN110835645AComprehensive detectionEasy to detectMicrobiological testing/measurementHigh Resolution Melt AnalysisBioinformatics
The invention relates to a digital PCR detection method, when the nucleic acid amplification reaction liquid to be detected is prepared, genotyping, mutation scanning, methylation research and the like can be realized by using dyes, high resolution and sensitivity are achieved, and the detection cost is reduced. Moreover, a polymerase chain reaction of a micro-droplet array can be completed on a same highly-integrated digital PCR detector, and melting curve analysis is carried out on a PCR product after PCR amplification of the micro-droplet array. Meanwhile, through the digital PCR detectionmethod, a fluorescence curve and a melting curve of the micro-droplet array can be obtained, and traceless connection between real-time monitoring of the whole PCR amplification process and melting curve analysis of the PCR product can be completely achieved. Therefore, copy number identification based on different fluorescence curves, identification of nucleic acid amplification characteristics based on different characteristic melting curves and typing, mutation scanning and the like of genes through high-resolution melting curves are realized, and digital PCR detection is completed more comprehensively, simply, conveniently and efficiently.
Owner:思纳福(苏州)生命科技有限公司
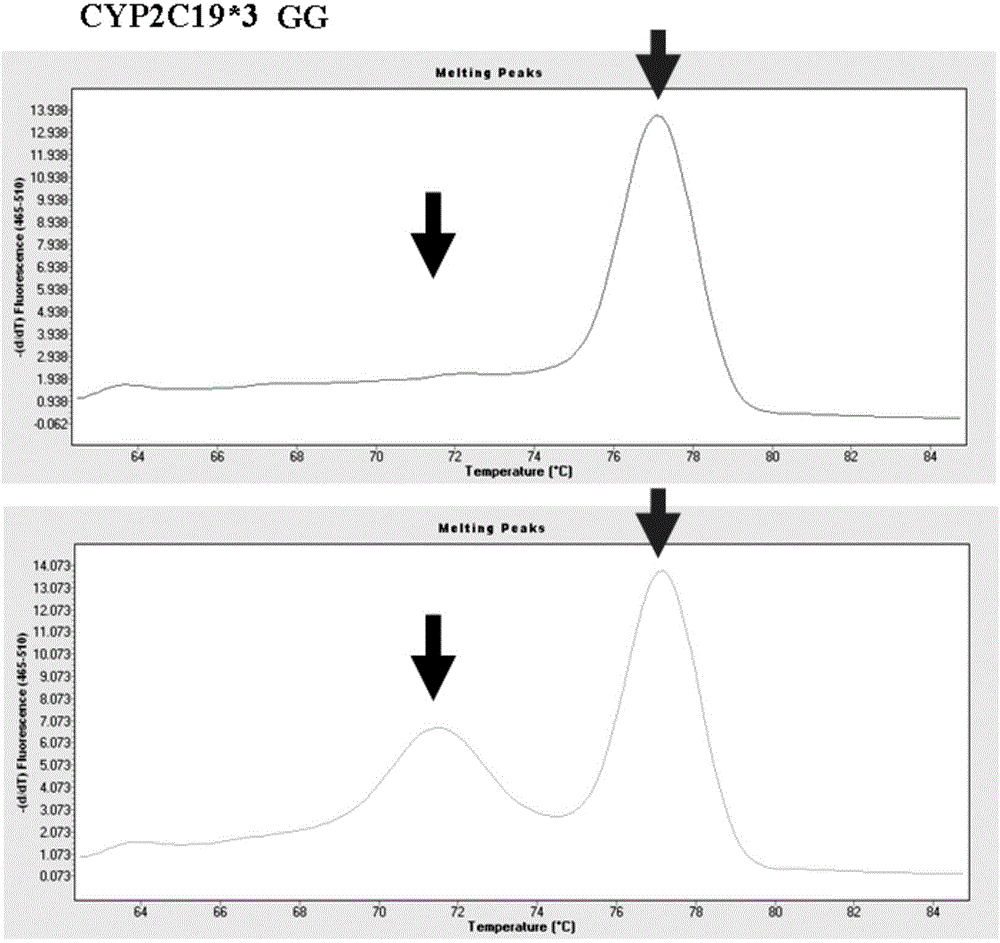
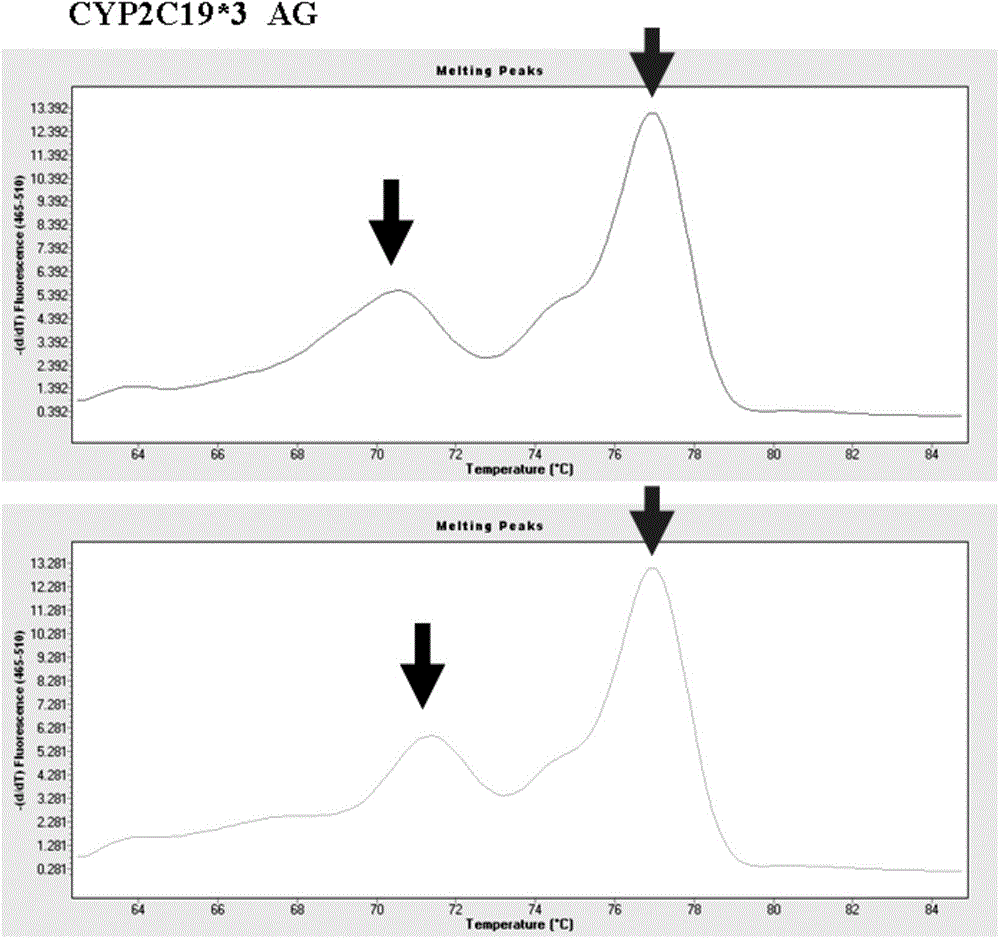
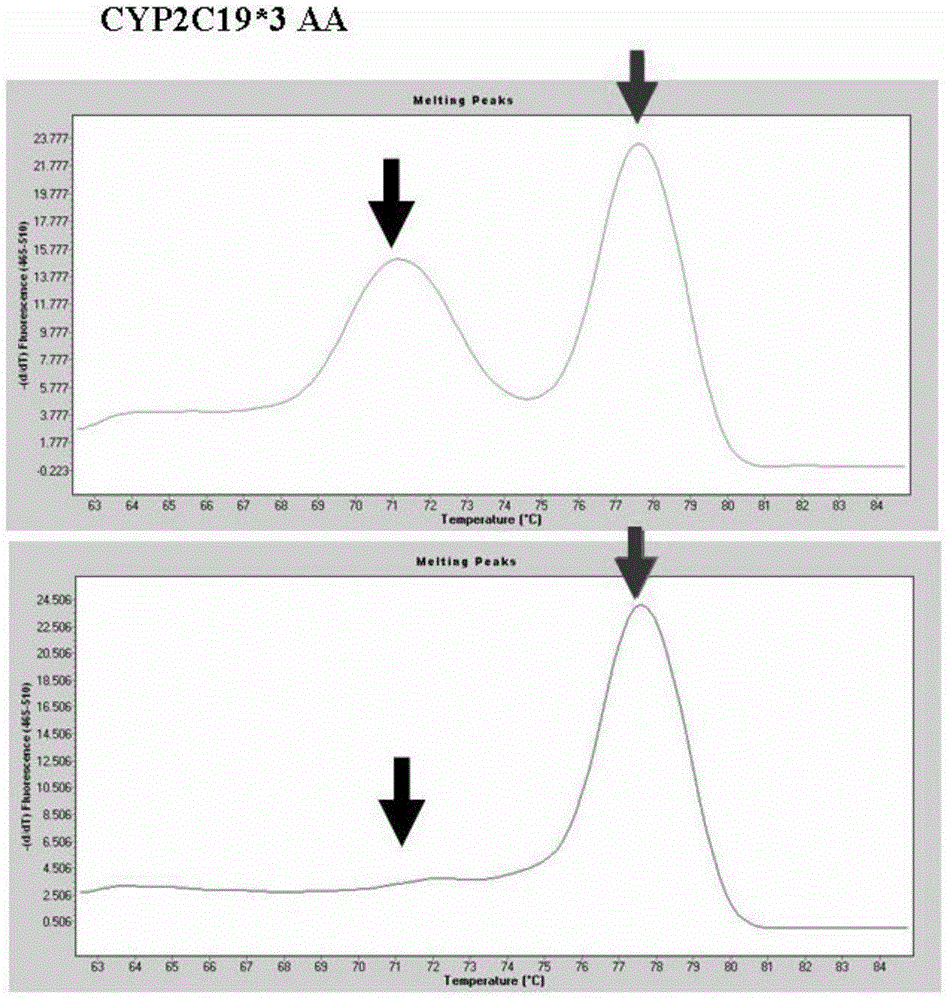
Primer combination for detecting allele CYP2C19*3 and detection kit thereof
ActiveCN103952481ASimple methodLow costMicrobiological testing/measurementDNA/RNA fragmentationFluorescencePrimer extension
The invention provides a primer combination for detecting an allele CYP2C19*3. The primer combination includes an upstream primer p1, a downstream primer p2, a gene extension primer p3 and a gene extension primer p4 respectively having the sequences shown in Seq ID No.1-4. The invention also provides a primer combination-containing kit for detecting the allele CYP2C19*3. Based on design thoughts of specific primer extension and a high-resolution melting curve, genotyping can be directly carried out in a premise of not carrying out special amplification post-processing. The method perform genotyping by using a fluorescent dye identification extension product, requires no use of special labeled probes, and is simple in method, low in cost, and easy to popularize in clinic.
Owner:BEIJING ERRUI XINYUE TECH CO LTD
Detection method and application of rice blast resistance gene Pigm
ActiveCN106929585AConvenienceSensitivityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGenome Scan
The invention relates to a detection method and application of a rice blast resistance gene Pigm, belongs to the technical field of agricultural biology and particularly relates to a specific molecular marker for High-Resolution Melting Curve Analysis (HRM) of the rice blast resistance gene Pigm and a method for detecting the rice blast resistance gene Pigm by virtue of a primer. According to the detection method, a rice 60K gene chip is utilized for carrying out whole-genome scanning on Gumei #4, and wuyunjing #7 and 9311, an HRM primer is designed by virtue of different variation sites, on which Gumei #4 are different from wuyunjing #7 and 9311, close to two ends of the gene Pigm, a molecular marker completely co-separated from Gumei is developed at each of two ends, and a middle-high-flux assistant selection system is established, so that the disease-resistant breeding efficiency of rice can be greatly improved.
Owner:SHENZHEN XINGWANG BIOLOGICAL SEED IND +1
Method for detecting drug-resistant mutation sites of campylobacter jejuni carbostyril antibiotics
ActiveCN103695559ASimplify complex proceduresGood sensitivity and specificityMicrobiological testing/measurementMicroorganism based processesDrugCampylobacter jejuni
The invention discloses a method for detecting drug-resistant mutation sites of campylobacter jejuni carbostyril antibiotics. The method comprises the following steps: by taking campylobacter jejuni of which the 257th site of a gyrA gene coding sequence is C as a wild type standard strain, taking campylobacter jejuni of which the 257th site of the gyrA gene coding sequence is T as a mutant type standard strain, respectively and simultaneously performing high-resolution melting curve analysis with campylobacter jejuni to be detected; determining whether the 257th site of the gyrA gene coding sequence of the campylobacter jejuni to be detected is C or T according to the comparison result of the high-resolution melting curve; if the high-resolution melting curve of the campylobacter jejuni to be detected is consistent with that of the wild type standard strain, selecting the campylobacter jejuni to be detected as a strain of which the 257th site of the gyrA gene coding sequence is C, and if the high-resolution melting curve of the campylobacter jejuni to be detected is consistent with that of the mutant type standard strain, selecting the campylobacter jejuni to be detected as a strain of which the 257th site of the gyrA gene coding sequence is T.
Owner:CHINA AGRI UNIV
Combination primer for human gall stone related gene mutation screening and application thereof
InactiveCN108060214AImprove throughputLow costMicrobiological testing/measurementDNA/RNA fragmentationForward primerHigh flux
The invention discloses a combination primer for human gall stone related gene mutation screening and application thereof, and relates to the technical field of gene detection. The combination primerfor the human gall stone related gene mutation screening is divided into a combination forward primer and a combination reverse primer; a human gall stone related gene mutation screening kit comprisesthe combination primer and LightScanner Master Mix. The invention provides a human gall stone related gene mutation screening method; the method is based on a high-resolution melting curve method; three mutation sites of common gall stone related gene can be simultaneously screened; common mutation sites such as rs1260326, rs1256049 and rs11887534 of related genes on chr2 and chr14 chromosomes are included. The characteristics of high flux, low cost and short detection time are realized.
Owner:深圳市一道生物科技有限公司
Methods for DNA targets detection directly in crude samples through polymerase chain reaction and genotyping via high resolution melting analysis
InactiveCN110945142AEasy self collectionInnovative multiple technologiesMicrobiological testing/measurementChemical compoundSingle strand
A method id disclosed for DNA targets detection directly in crude samples through Polymerase Chain Reaction (PCR) and genotyping via High Resolution Melting (HRM) analysis. The method comprises diluting the crude sample, treating the diluted crude sample by performing a ramp of increasing and then decreasing temperature, performing PCR amplification and then an HRM analysis. The method further comprises monitoring, during the HRM analysis, the change in the signal emission resulting from the temperature-induced denaturation of the double-stranded amplicons into two single- stranded DNA, due tothe release of an intercalating molecule or compound. The detection of DNA targets in the crude sample is performed through a reader analysing the signal variation, obtaining the result of the analysis through a graphic interface connected to the reader.
Owner:ULISSE BIOMED
In-vitro detection method for quickly screening exon 5-8 mutation of P53 gene and application
InactiveCN106834487ALow priceLow costMicrobiological testing/measurementDNA/RNA fragmentationExonGene mutation
The invention provides an in-vitro detection method for quickly screening the exon 5-8 mutation of a P53 gene. The method comprises the following steps: (1) extracting the genome DNA of a peripheral blood sample; (2) designing and synthesizing 4 pairs of specific primers for amplifying exon 5-8 of the P53 gene; (3) performing high resolution melting curve analysis and grounding PCR on the genome DNA of the sample by using the primers; and (4) obtaining the sample with a positive result in the curve analysis, meaning occurrence of P53 gene mutation. The detection method is not limited by site or type of mutated base, does not need a sequence specific probe, and can directly run high resolution melting after PCR is completed to complete analysis on the sample mutation. The method is simple, convenient and quick in operation, is low in use cost and accurate in result, can realize truly closed pipe operation. Furthermore, the method has the characteristics of high through-put, high speed, kit availability, low detection cost and the like for quick screening of population.
Owner:北京青航基因科技有限公司
High-resolution melting analysis
InactiveUS8180572B2Heating or cooling apparatusSugar derivativesHigh Resolution Melt AnalysisComputational biology
The present invention relates to methods and systems for the analysis of the dissociation behavior of nucleic acids and the identification of nucleic acids. In one aspect, methods and systems are disclosed for resolving a denaturation curve of a sample containing a first and second nucleic acid into a resolved denaturation curve for the first nucleic acid and a resolved denuration curve for the second nucleic acid.
Owner:CANON US LIFE SCIENCES INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com