High resolution melting curve-based multi-SNP identification method
A high-resolution melting and identification method technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of high requirements, poor stability, and limited HRM analysis efficiency, so as to reduce experimental costs and improve expansion. The effect of increasing fragment specificity and improving experimental efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
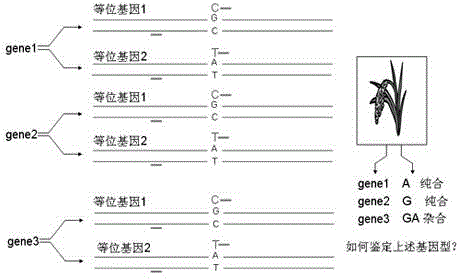
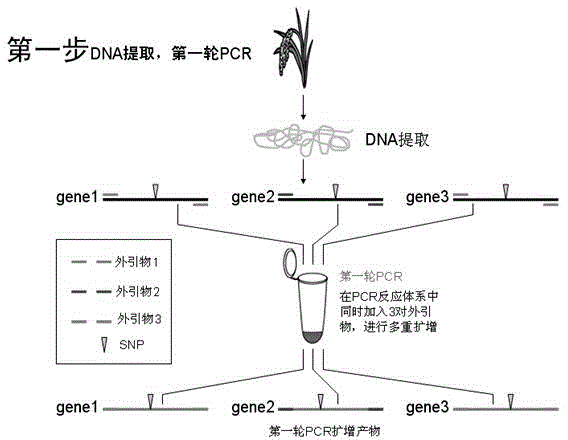
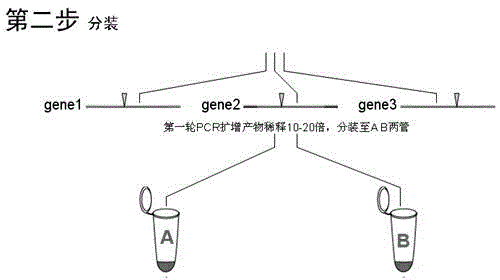
[0062] A rice material "Texianzhan 13" identified by HRM technology Wx Pita Pik genotype method
[0063] 1. Extraction of Genomic DNA from Special Indica Zhan 13:
[0064] Specific methods: (1) Take a small amount of one-month-old rice leaves after transplanting, put them in a 2.0 mL sterilized centrifuge tube frozen in liquid nitrogen, stir until powdery, add 1000 μL 2×CTAB-DNA extraction solution (mass fraction W / V 2% CTAB, pH8.0; mass fraction W / V 1% PVP; 100 mmol / L Tris-HCl, pH8.0; 1.4 mol / L NaCl; 20 mmol / L EDTA, pH8.0; volume fraction V / V 0.2% mercaptoethanol); (2) placed in a constant temperature water bath at 65 °C and shaken every 10 min, and removed after 30-45 min; (3) after cooling for 2 min, add 1000 μL of chloroform with a volume ratio of 24:1- Shake isoamyl alcohol vigorously up and down to mix the two evenly; (4) Centrifuge at 10,000 rpm for 10 min, gently remove the supernatant into a 1.5 ml sterilized new centrifuge tube, add 600 μL of pre-cooled isopro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com