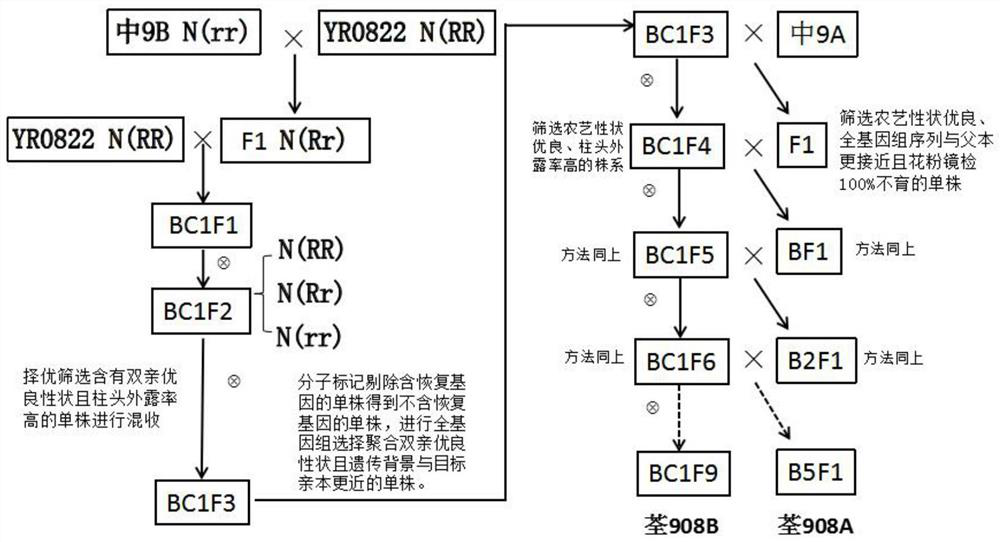
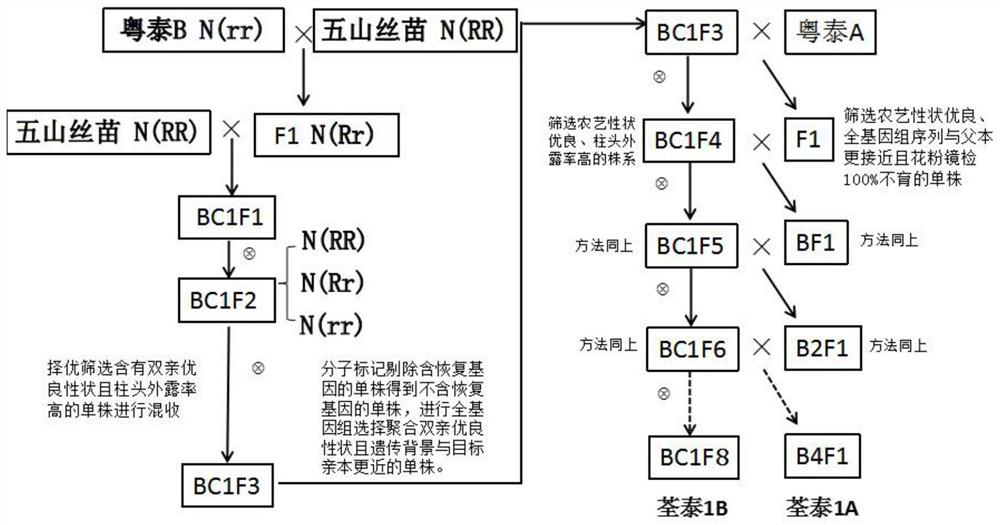
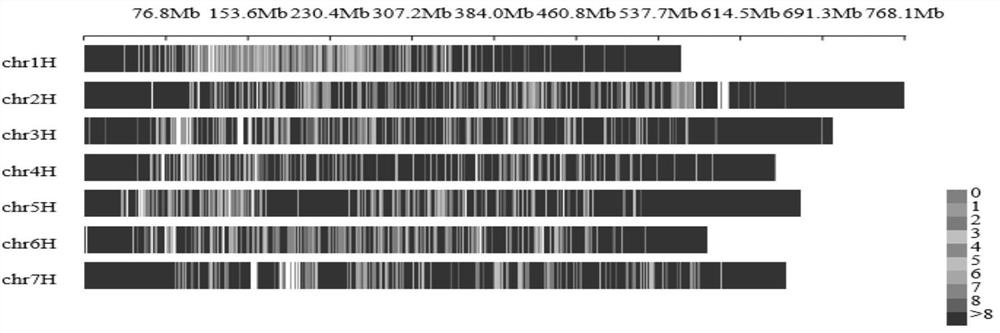
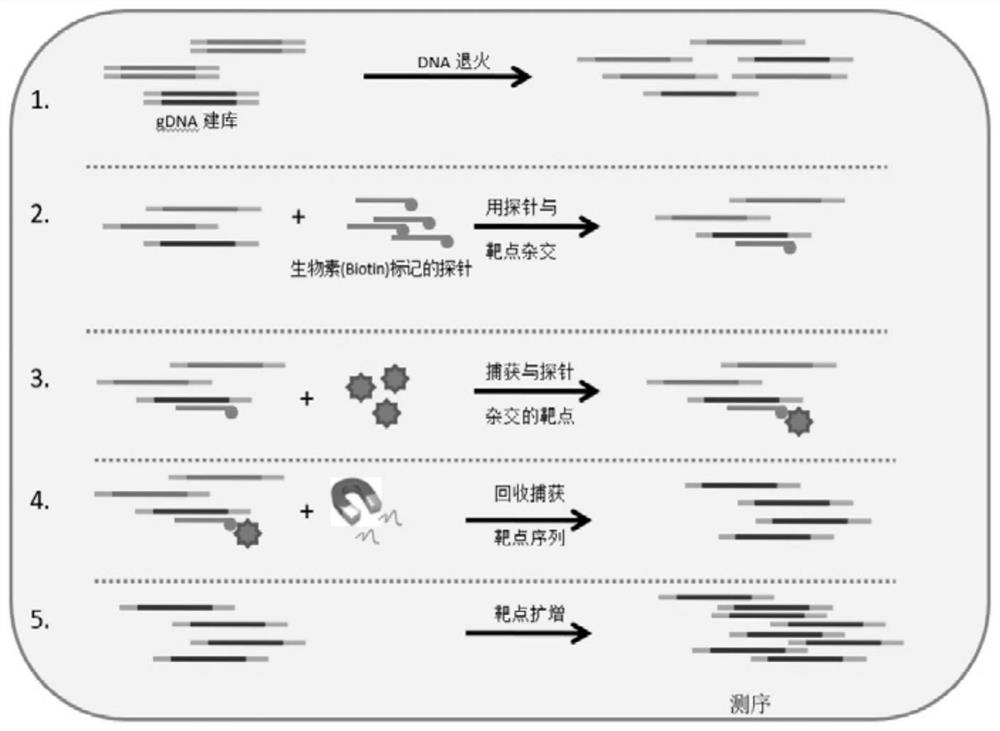
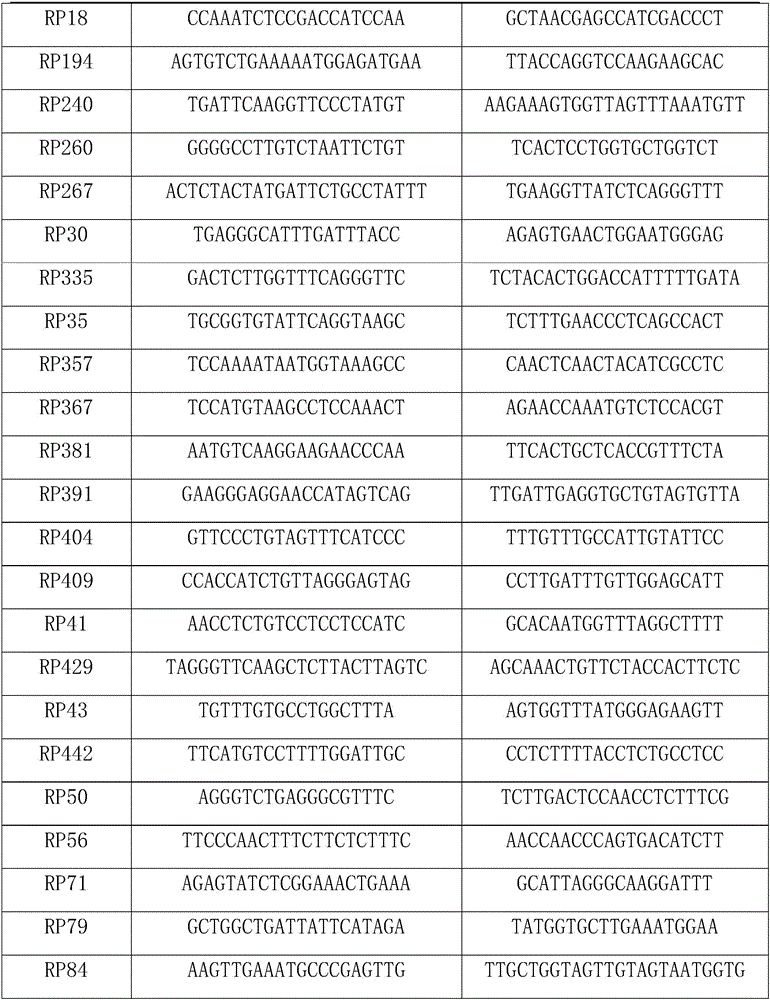
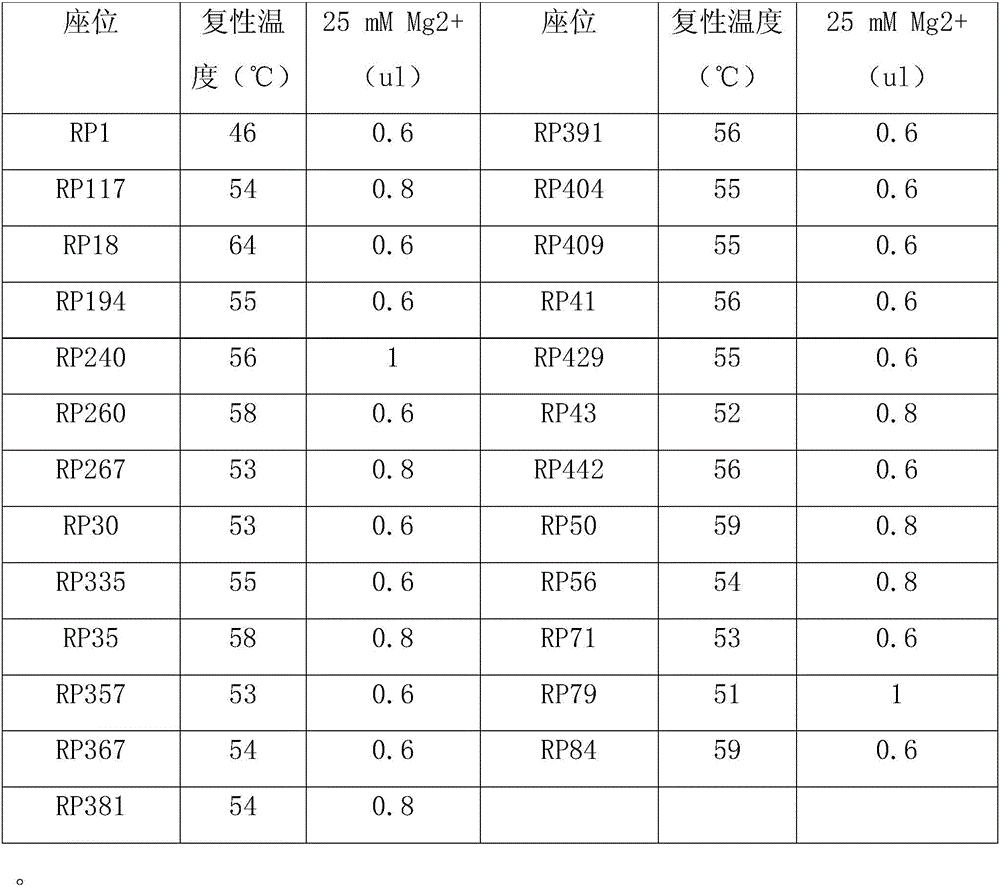
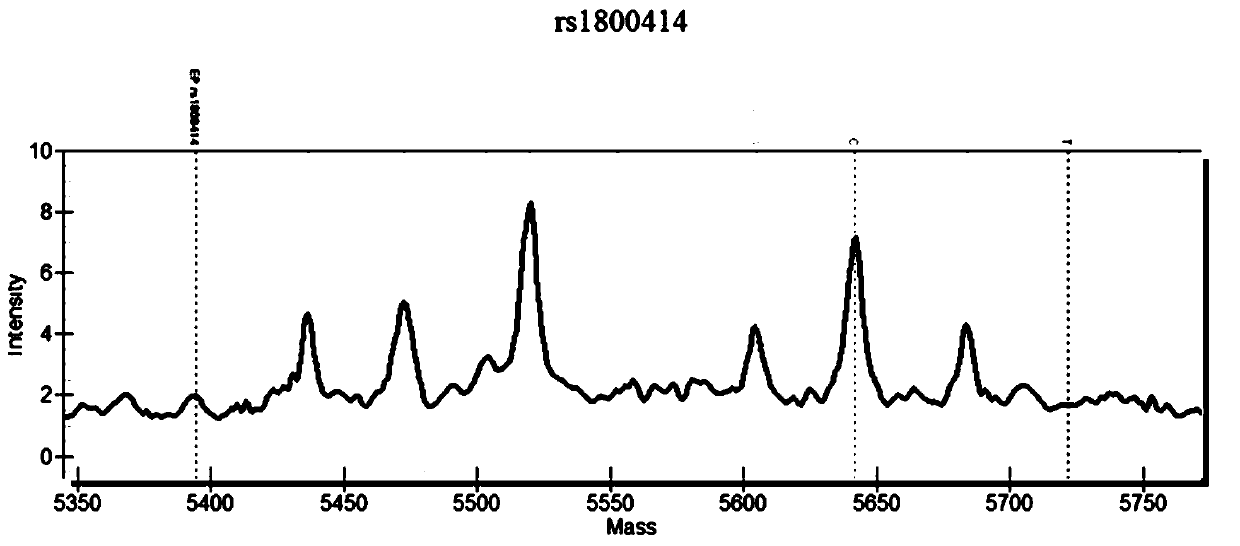
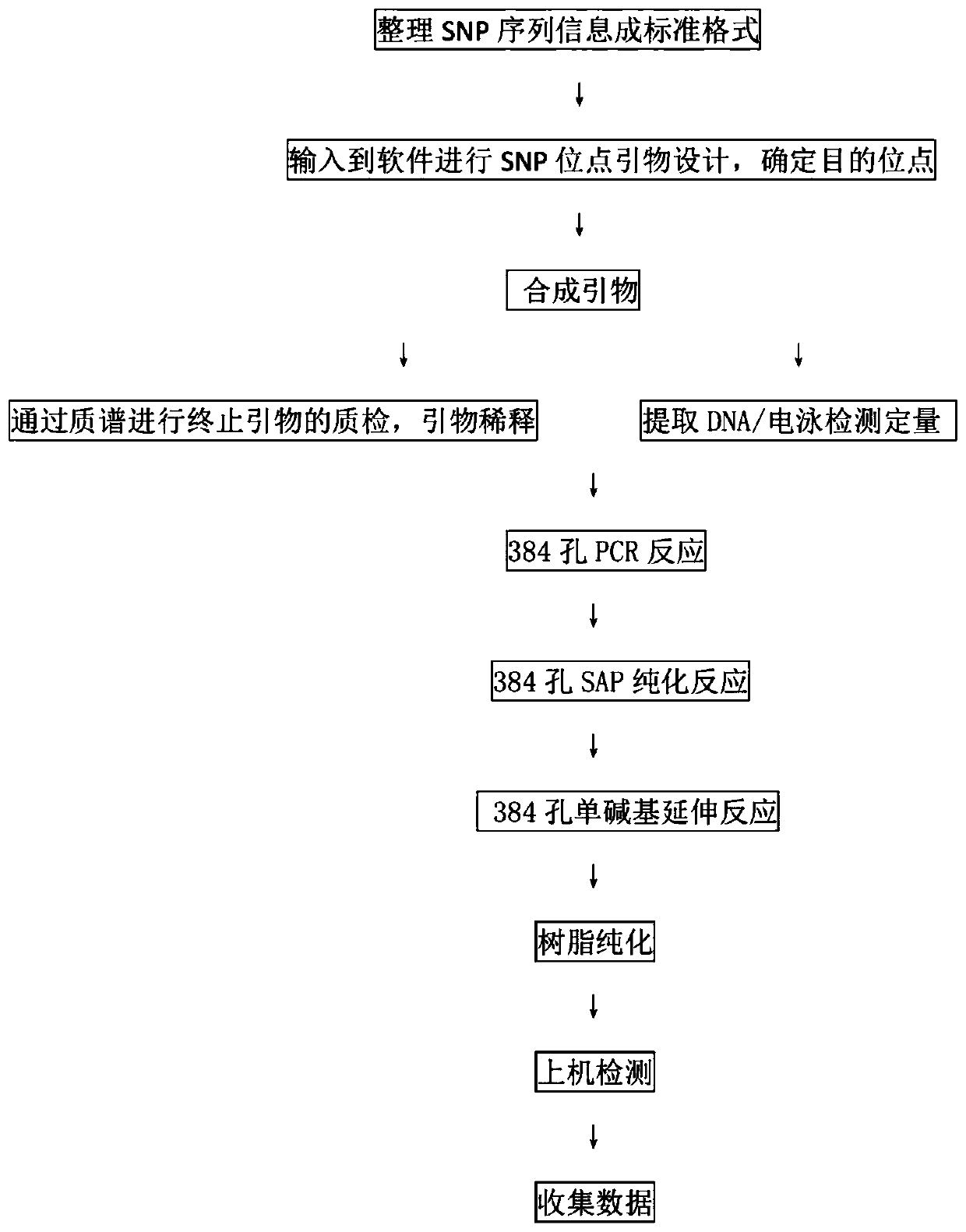
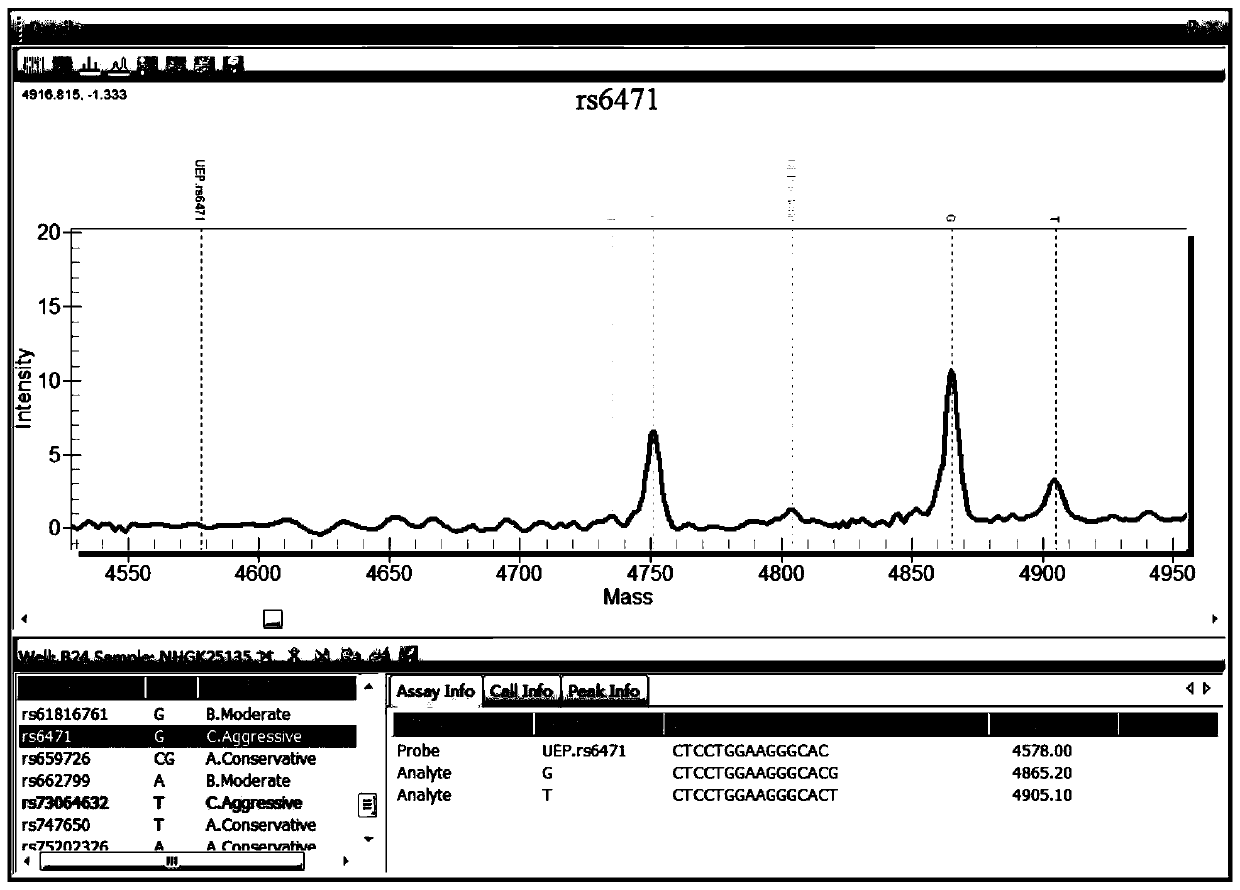
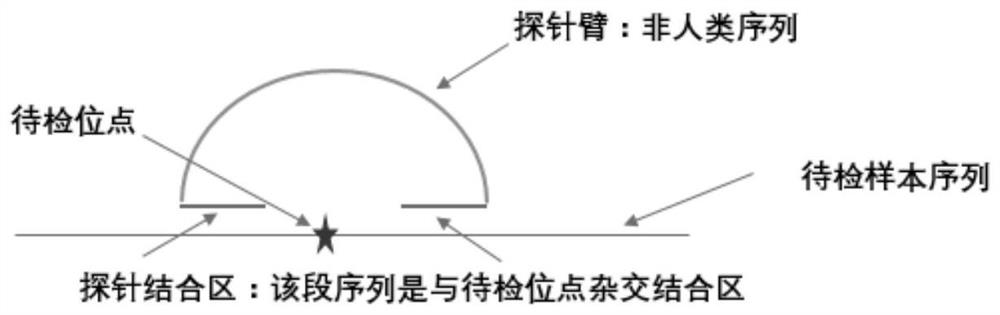
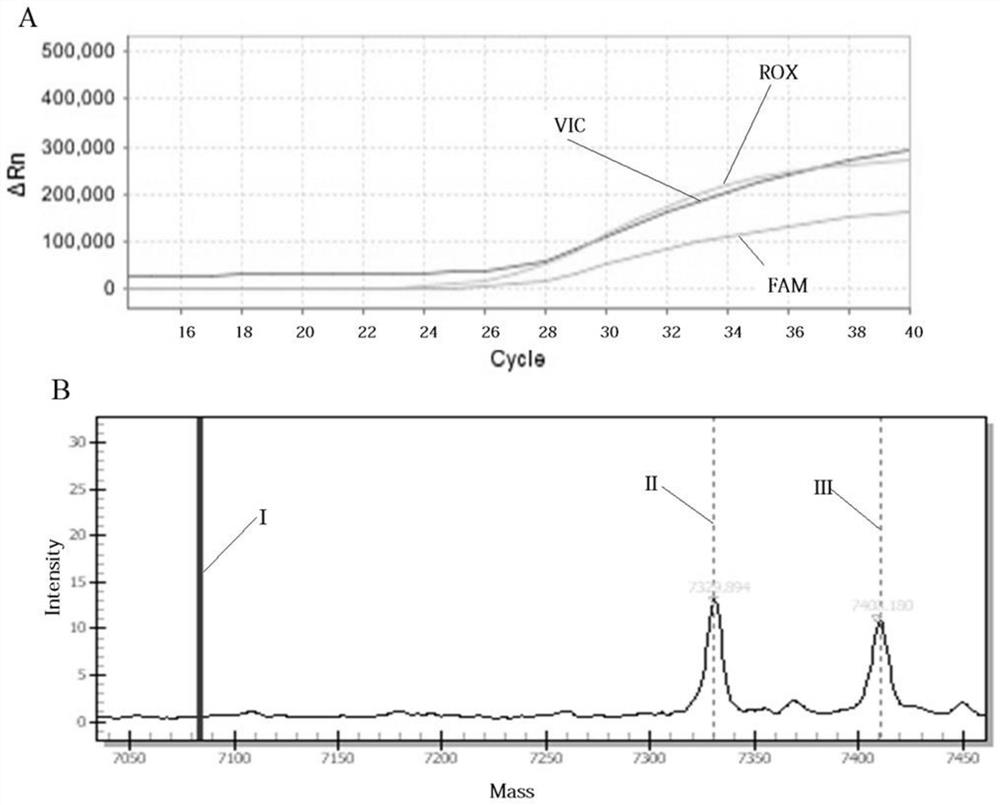
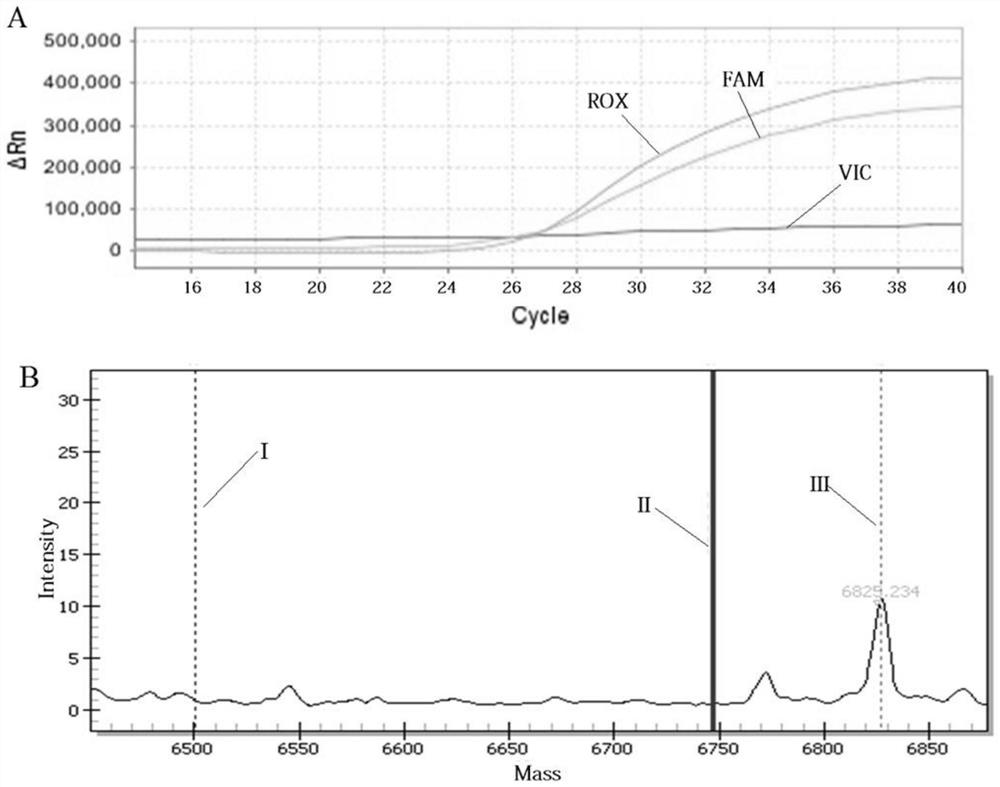
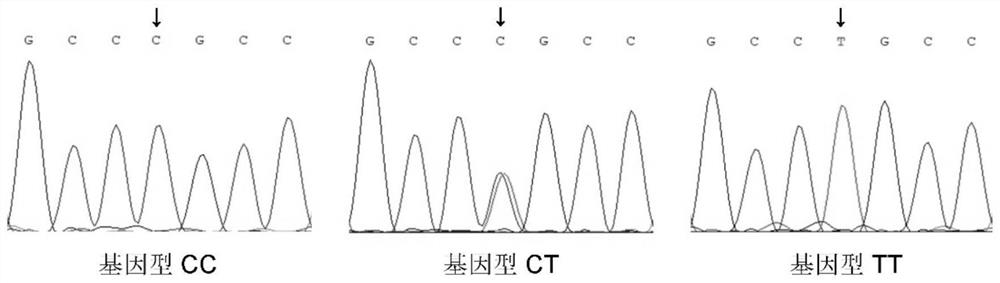
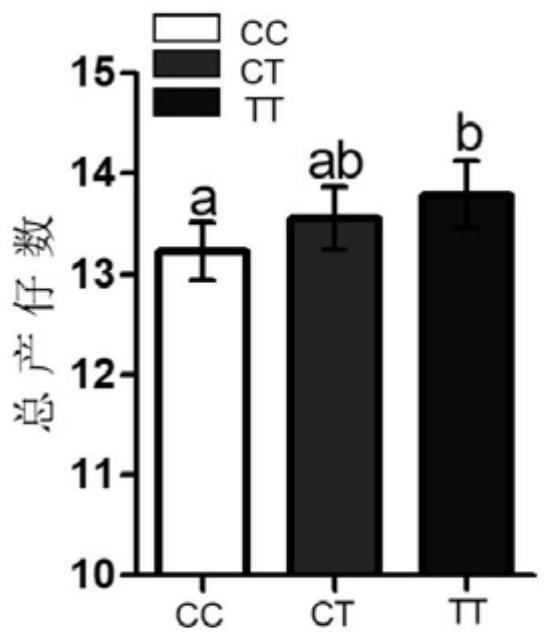
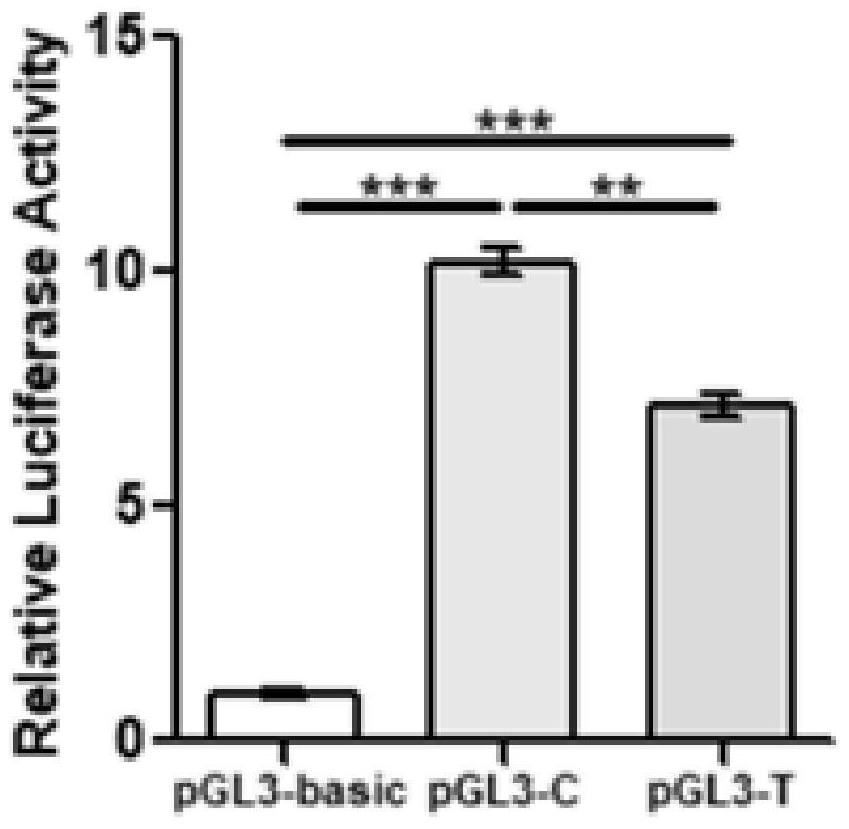
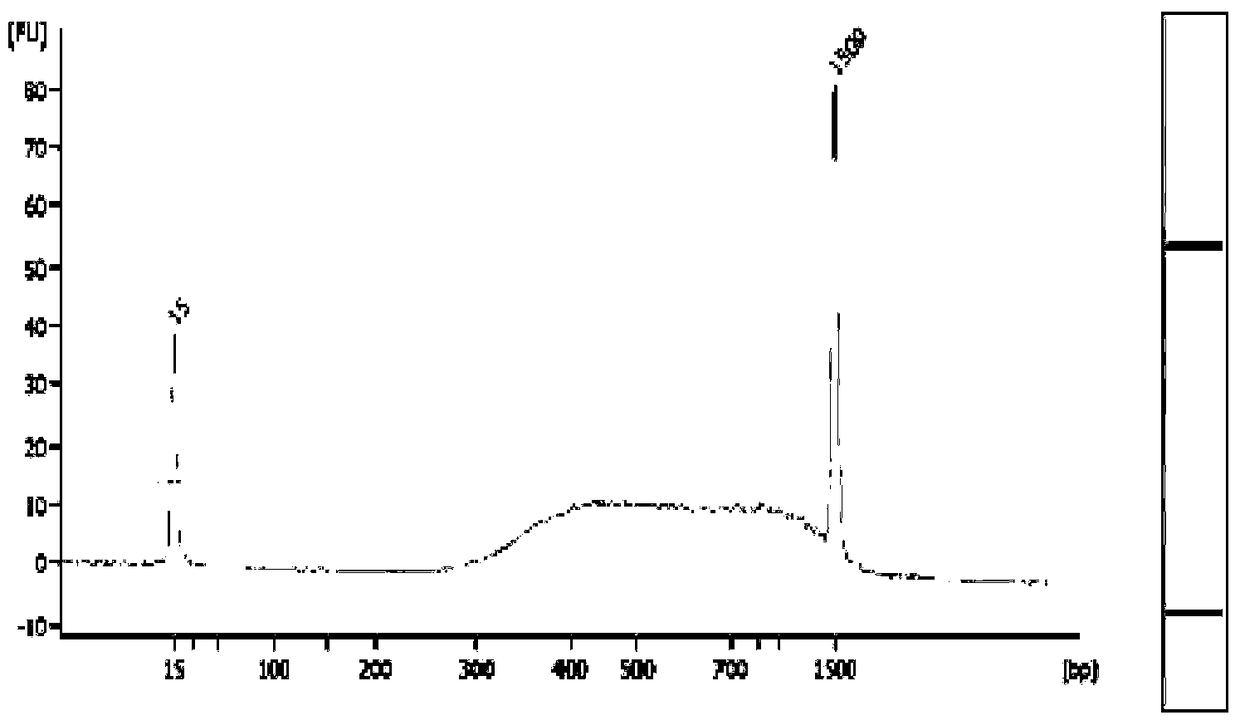
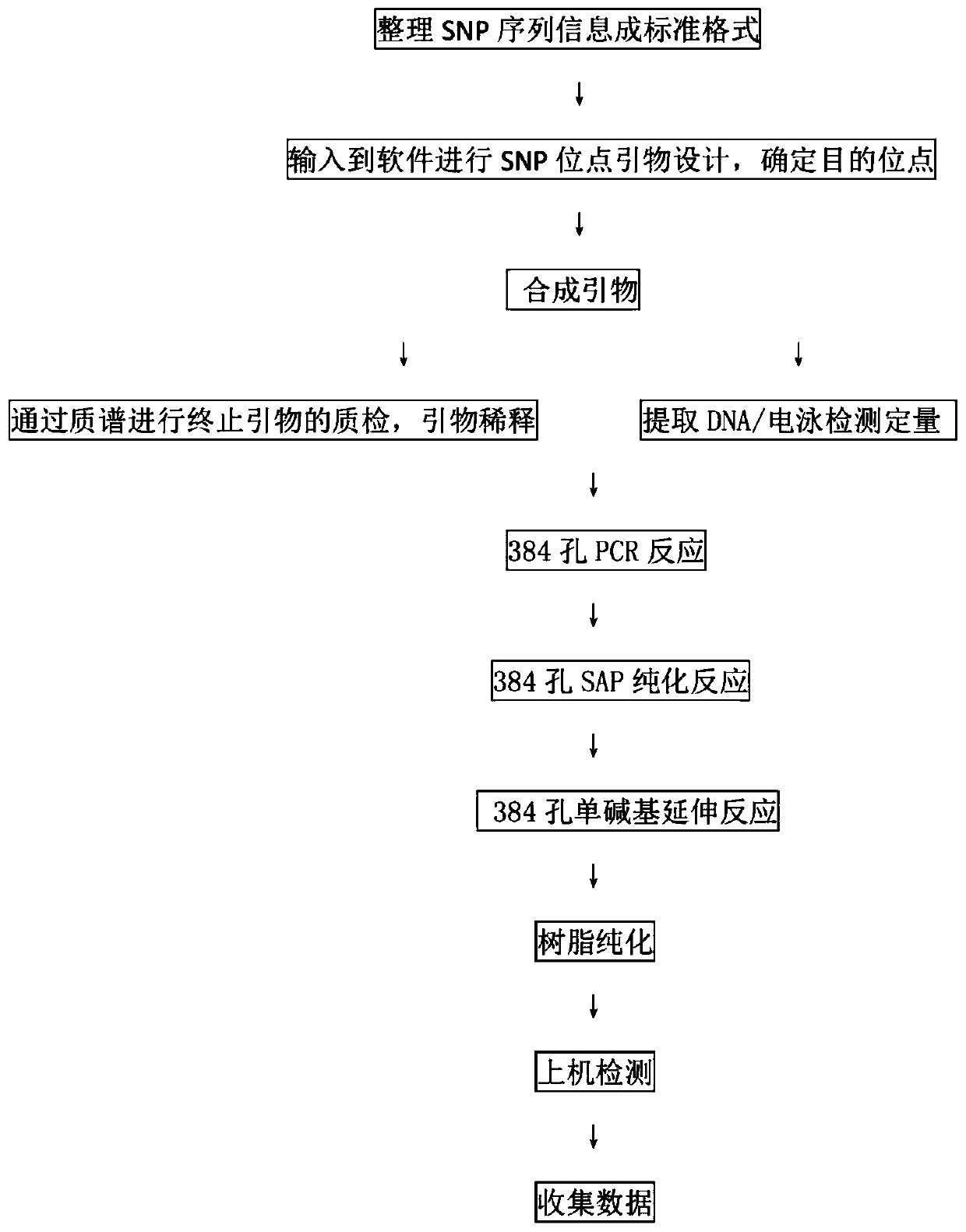
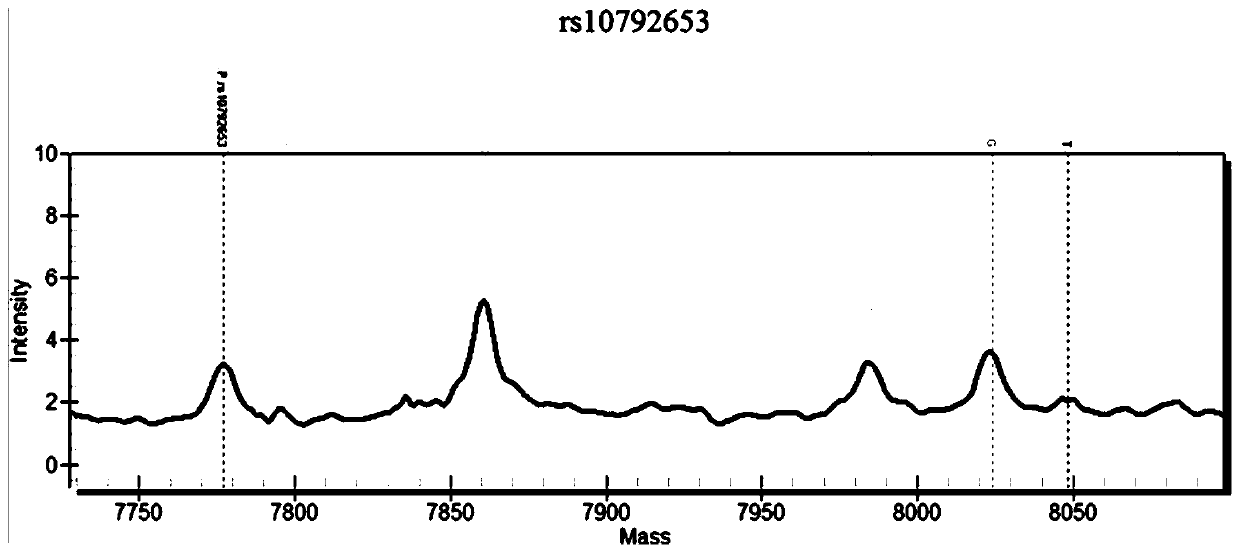
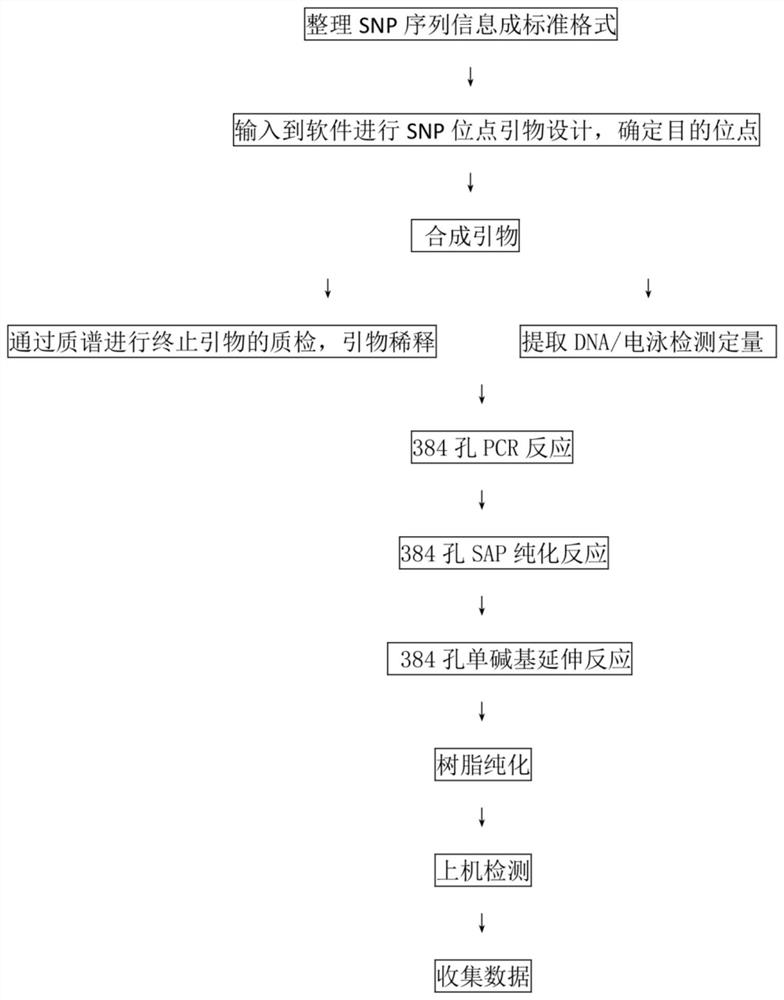
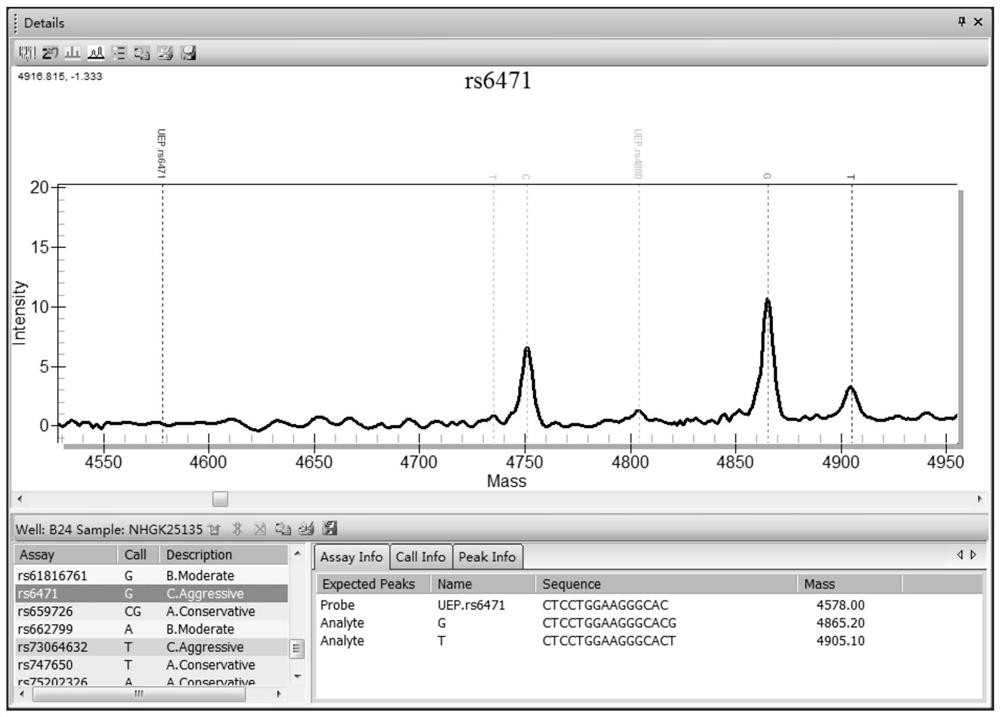
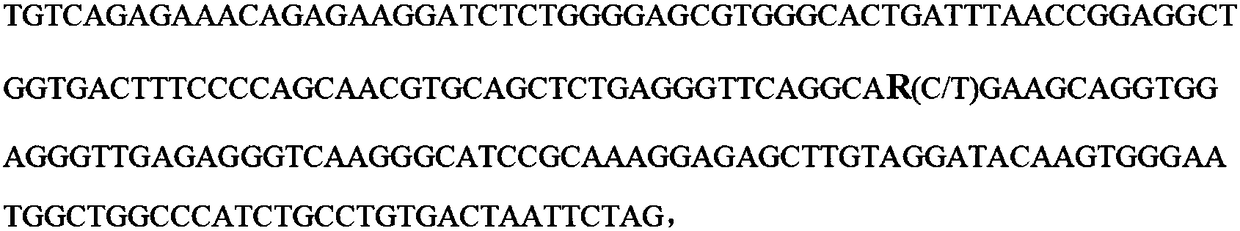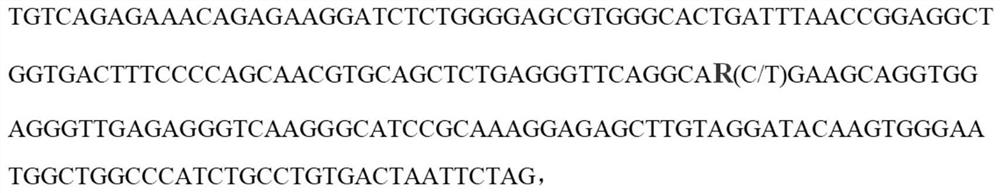Patents
Literature
34 results about "Genotyping Techniques" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Methods used to determine individuals' specific ALLELES or SNPS (single nucleotide polymorphisms).
Fluorescence quantitative detecting method for CYP2C19 genotyping
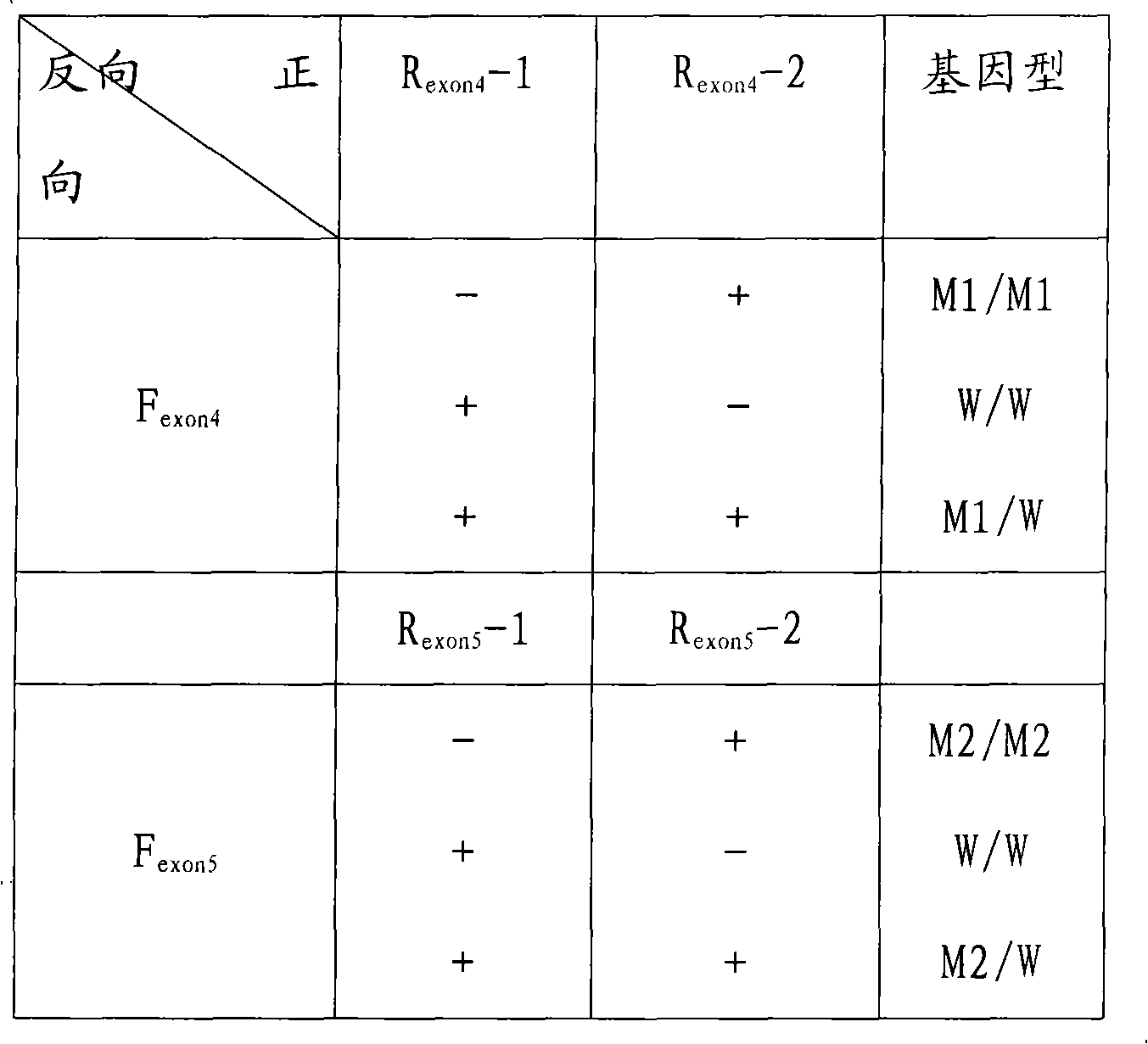
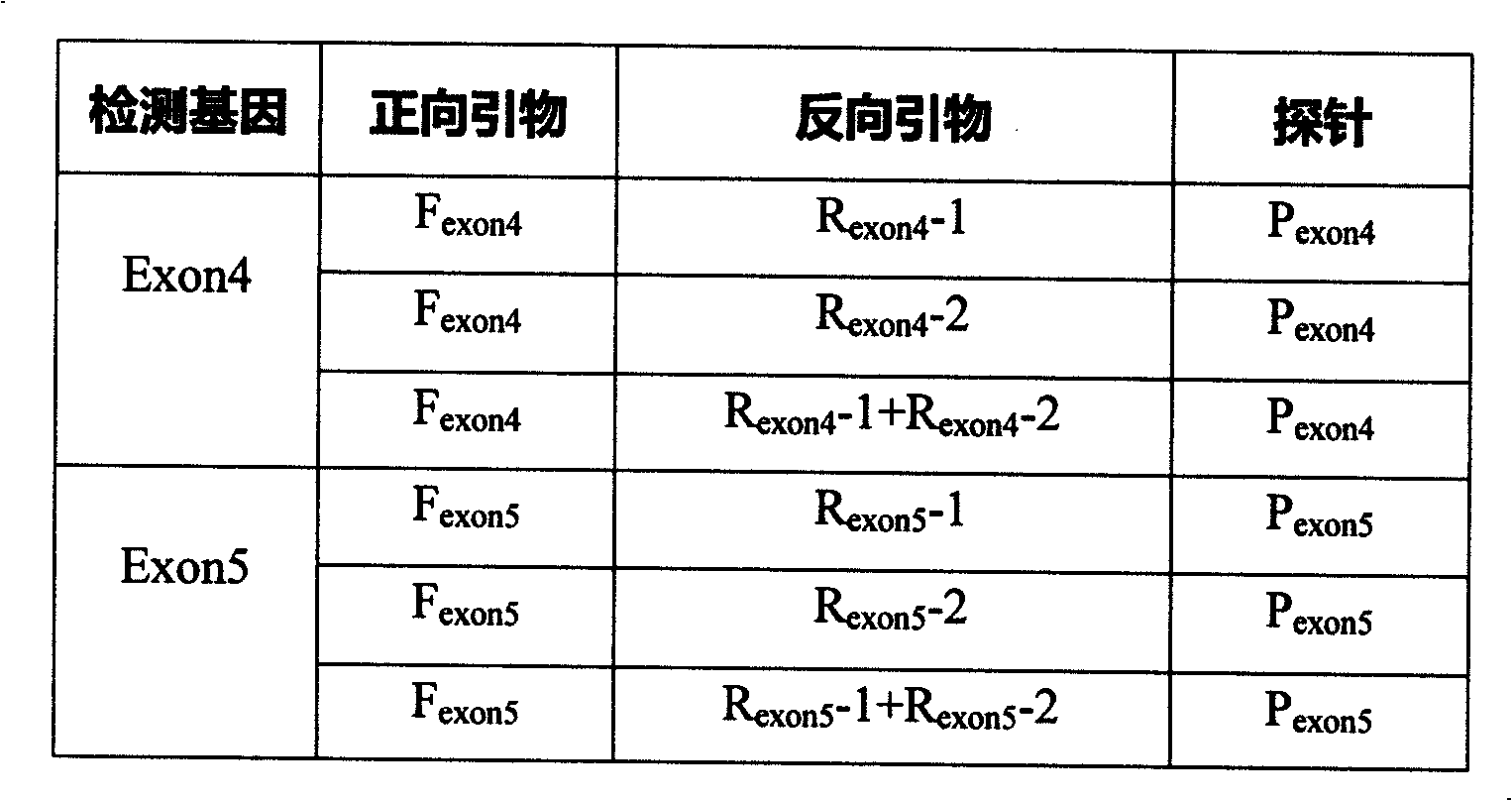
The invention relates to a fluorescent quantitative detecting method for CYP2C19 genotyping fluorescent and a diagnostic kit. At present, CYP2C19 genotyping techniques at home and abroad are mainly RFLP technique and ALA technique which determine genotypes according to the size of a DNA fragment, with the disadvantages of time-consuming operation and low flux. The invention provides a novel point mutating or SNP detecting method which utilizes the ASA combination primer sequence designed for the polymorphic loci of the CYP2C19 gene exons 4 and 5, the referential ASA primer combination or the degenerated primer sequence for quality control, specific TaqMan-MGB probe sequences for amplified products, an ASA amplifying reaction method, the amplifying reaction result of real-time fluorescent quantitative detection, the fast analysis of mutation locus type and genotyping. The quantitative detecting method has the advantages of time saving compared with a conventional ASA method, no need for electrophoresis detection, fastness, accuracy, and the like, and can be used for detecting other drug-metabolizing enzymes, or more extensive genetic variation or mutation.
Owner:樊世斌 +1
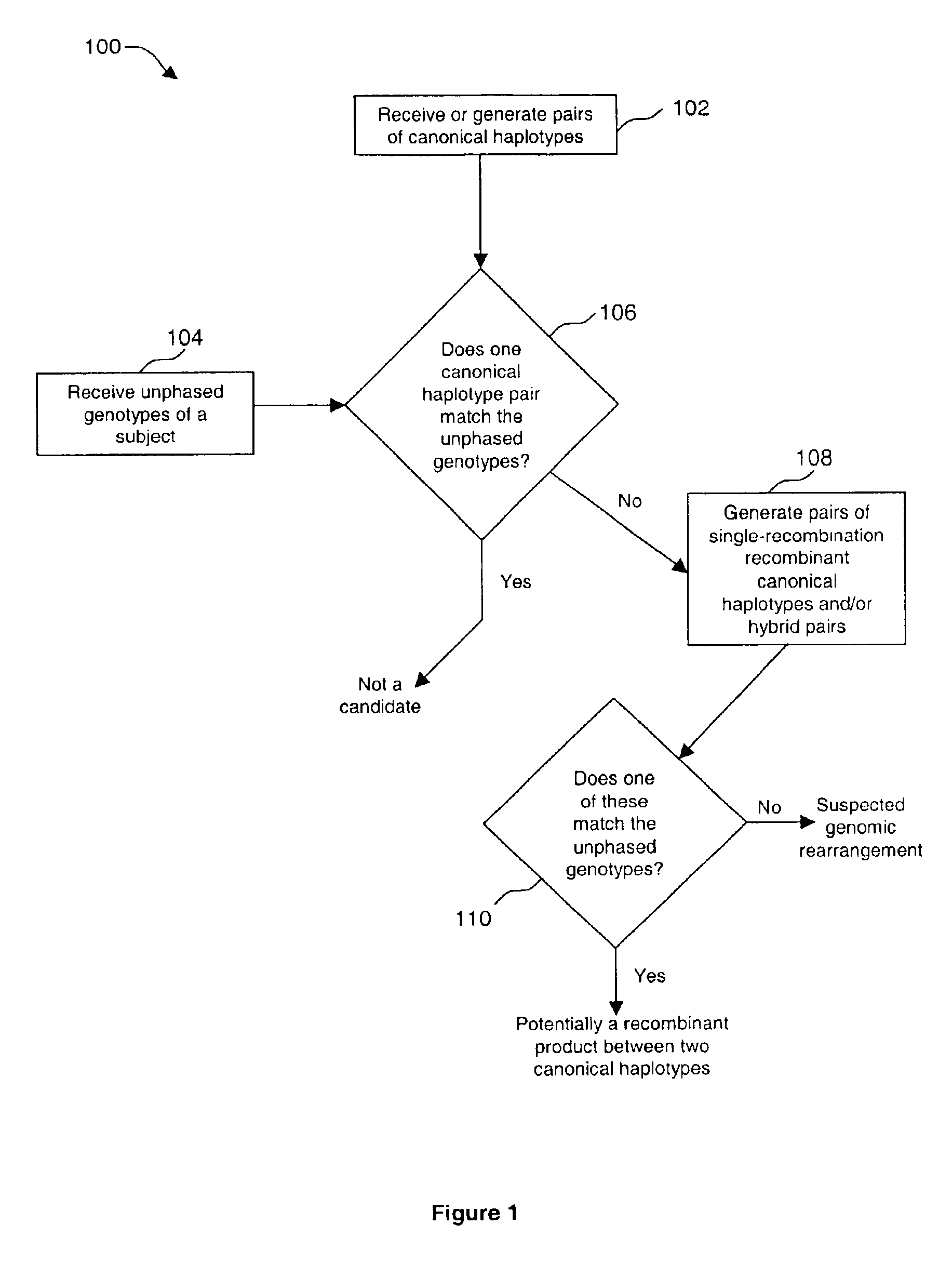
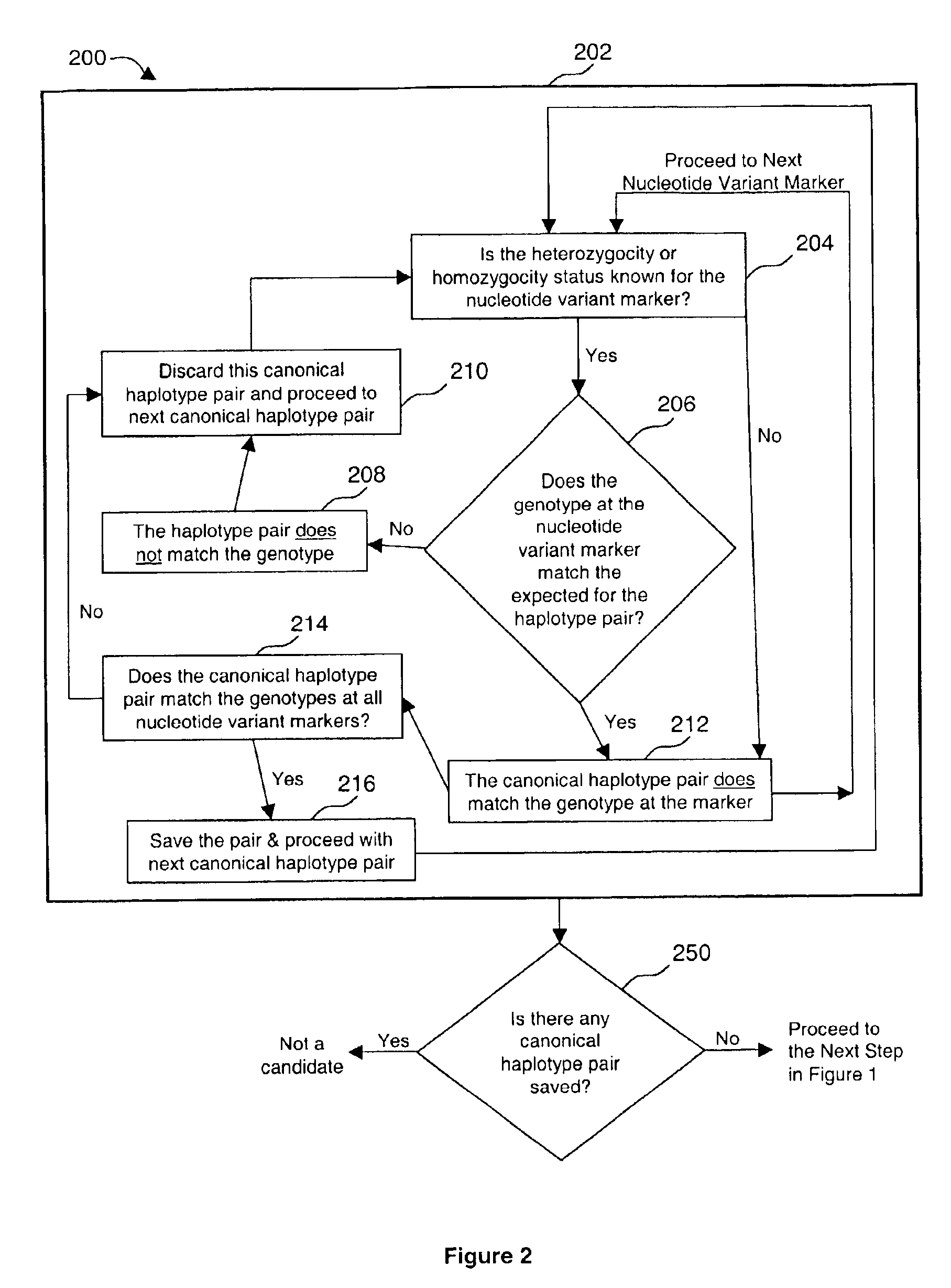
Method of identifying genomic rearrangements
InactiveUS6895337B1Quickly and accurately analyzedImprove accuracyComputer controlRead-only memoriesHeterologousHaplotype
Methods, computer program products and systems are provided for detecting large genomic rearrangements based on unphased genotype data obtained using common genotyping techniques that do not differentiate different alleles. In the method, unphased genotypes at a plurality of nucleotide variant markers of a particular gene in a diploid subject are compared with a canonical haplotype collection of the gene for a heterogeneous subject population. If the unphased genotypes cannot be reduced to a pair of canonical haplotypes within the canonical haplotype collection, it would indicate an increased likelihood that an allele of the gene in the diploid subject harbors a genomic rearrangement.
Owner:MYRIAD GENETICS
Determining method for sequencing enzyme digestion combination in sequencing genotyping technology
ActiveCN105368930AGuaranteed accuracyLow parting costMicrobiological testing/measurementEnzyme digestionSpecific enzyme
The invention provides a determining method for sequencing enzyme digestion combination in a sequencing genotyping technology. The determining method comprises the following steps that 1, restriction enzyme digestion site predicting is performed on a target genome, and the number of enzyme digestion segments obtained through different enzyme digestion modes is counted; 2, a joint sequence and a PCR amplification primer sequence at the two ends of each enzyme digestion segment are designed according to the predicted enzyme digestion segments in the various enzyme digestion modes in the step 1; 3, sequencing libraries are constructed through a GBS technology for the different enzyme digestion modes; 4, sequencing is performed through the sequencing libraries constructed in the step 3; 5, SNP marker sites are obtained according to sequencing results; 6, the specific enzyme digestion combination for the target genome is determined according to the number of the SNP marker sites and the enzyme digestion segment sizes which are obtained through different enzyme digestion combinations.
Owner:CHINA AGRI UNIV
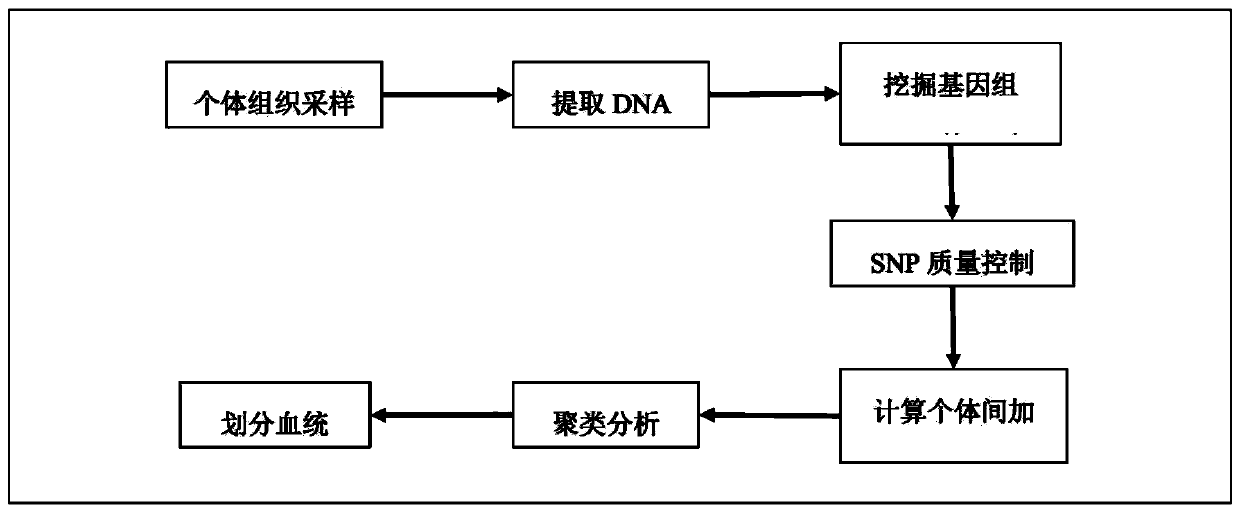
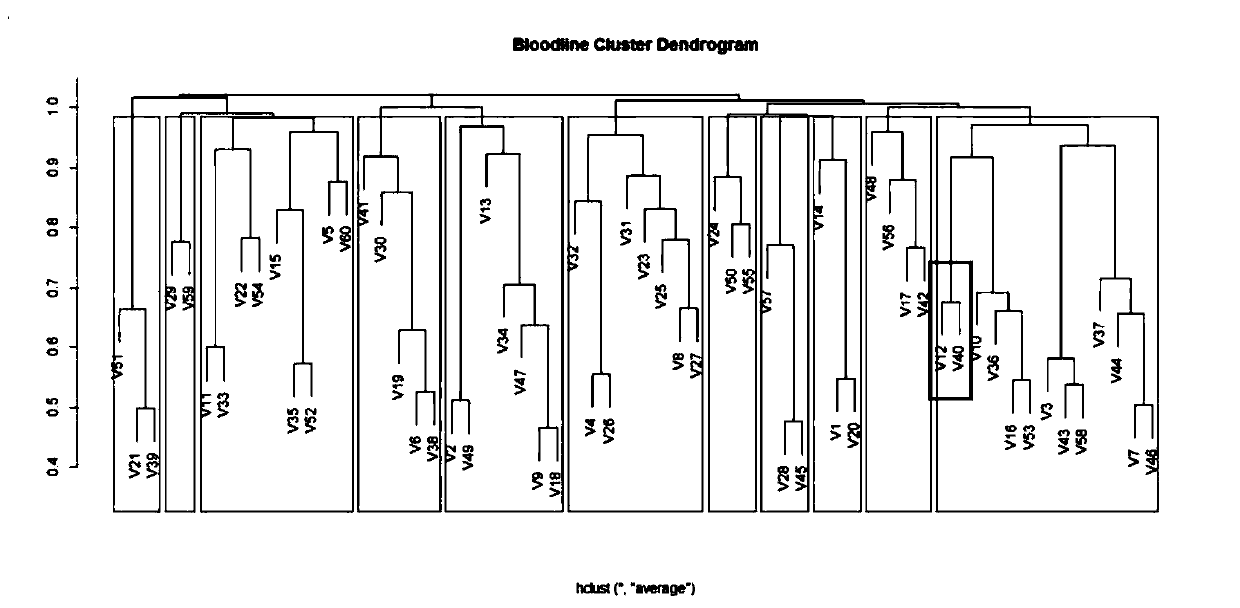
Method for dividing breeding swine blood linkage based on whole genome SNP information
ActiveCN110176274AAccurate divisionAccurate predictionMicrobiological testing/measurementProteomicsGenotyping TechniquesHeredity
The invention discloses a method for dividing breeding swine blood linkage based on whole genome SNP information. The method comprises the steps of acquiring a body tissue of a dividing object, extracting the DNA, typing the genome SNP of each individual through a certain gene typing technique, and screening a qualified SNP site through quality control. calculating an additive heredity correlationvalue between individuals through the genome SNP genotype, then subtracting the additive heredity correlation value from one and using the obtained value as the distance between two individuals, thenperforming cluster analysis on the two individuals through an R-language hclust function, and using 0.9844 as a threshold for dividing the blood linkage. Compared with a traditional blood linkage dividing method, the method according to the invention has advantages of realizing a clear and concise obtained result, sufficiently reserving allele information which is transmitted from the parents, and realizing high reliability in blood linkage dividing.
Owner:WENS FOOD GRP CO LTD +1
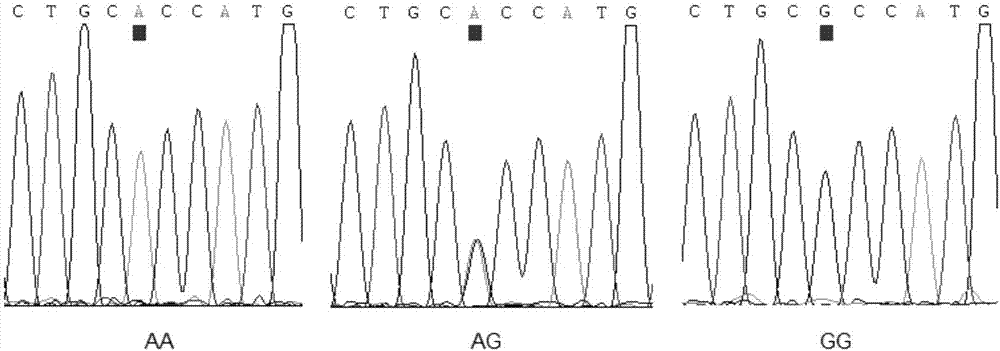
Gene mutation site related to reproduction traits of Hu-sheep and application thereof
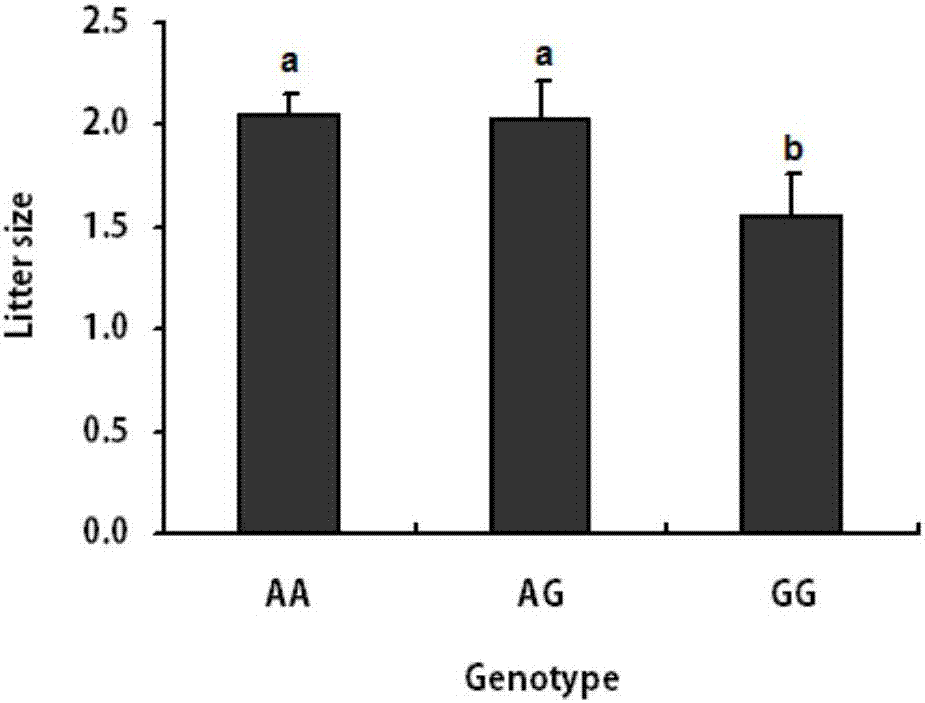
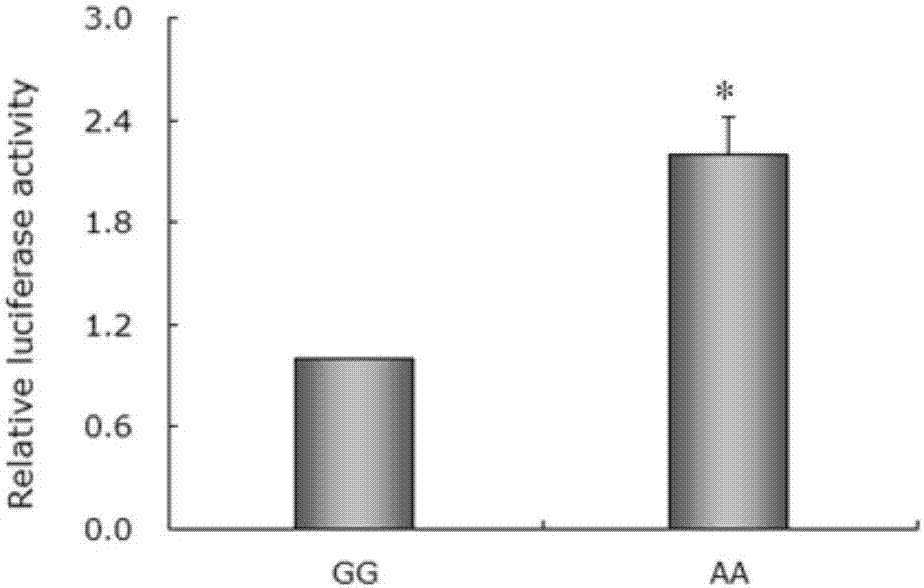
The invention discloses a gene mutation site related to reproduction traits of Hu-sheep and application thereof. The mutation site is located at Hu-sheep FSHR gene core promoter region-414 site nucleotide; A>G mutation is located at the site and is obviously related to the lambing quantity of the Hu-sheep, wherein AA and AG type individual lambing quantity is obviously higher than the GG type individual lambing quantity; the genetic transcription activity in AA type promoter region is obviously higher than that of GG type promoter region. The livestock reproduction trait is a low heritability trait; the genetic progress is slower when the conventional breeding technique is adopted; the mutation site is obviously related to the reproductive performance of Hu-sheep, so that the high-fecundity genotype individual is remained and the low-fecundity genotype individual is abandoned by adopting a simple genetic typing technique for detecting the site; the gene mutation site has an important economic value for quickening the breeding process of the high-fecundity Hu-sheep colony or strain.
Owner:NANJING AGRICULTURAL UNIVERSITY
Primer, probe, kit and method for detecting subtypes of human gene CYP1A2
InactiveCN106319073AAccurate detectionLow costMicrobiological testing/measurementDNA/RNA fragmentationForward primerNucleotide
The invention belongs to the technical field of genotyping and particularly relates to a primer, probe, kit and method for detecting subtypes of a human gene CYP1A2. The primer and the probe comprise the following nucleotide sequences: the nucleotide sequence of a forward primer is SEQ ID NO: 1: 5'-AAACTGAGATGATGTGTGGAGG-3'; the nucleotide sequence of a reverse primer is SEQ ID NO: 2: 5'-CACGCATCAGTGTTTATCAAA-3'; the nucleotide sequence of the probe is SEQ ID NO: 3: 5'-GTGGGCCCAGGACGCATGGTAGATGGA-3'. The kit comprises the primer, the probe, dNTPs, MgCl2, a mixed enzyme and positive and negative reference substances, wherein the mixed enzyme is prepared from a UNG enzyme and DNA polymerase with the activity of flap endonuclease. According to the method for detecting the subtypes of the gene CYP1A2 by using the kit, different subtypes of a target nucleic acid SNP locus can be detected by one fluorescent probe, so that the operating process is simplified, the cost is reduced, and the detection result is high in resolution and is rapid and accurate.
Owner:SHENZHEN UNI MEDICA TECH
Primer, kit and method for detecting human CYP2C9 genetic typing
InactiveCN106350595AAccurate detectionLow costMicrobiological testing/measurementDNA/RNA fragmentationNucleotideFluorescence
The invention belongs to the technical field of genetic typing, and particularly relates to a primer, a probe, a kit and a method for detecting human CYP2C9 genetic typing. The primer and the probe comprise the following nucleotide sequences: an upstream primer SEQ ID NO.1: 5'-CAGCTAAAGTCCAGGAAGAGAT-3', a downstream primer SEQ ID NO.2: 5'-TTCTGAATTTAATGTCACAGGT-3', and a probe SEQ ID NO.3: 5'-CAGAGATACATTGACCTTCTCCCCACC-3'. The kit comprises the primer and the probe, dNTPs (Deoxyribonucleoside Triphosphate), MgCl2, mixed enzyme prepared from UNG (Uracil-N-Glycosylase) and DNA (Deoxyribose Nucleic Acid) polymerase having the activity of flap endonuclease, and positive and negative reference substances. The method for detecting the human CYP2C9 genetic typing by using the kit is capable of detecting different types on a target nucleic acid SNP (Single Nucleotide Polymorphism) site, so that the operation process is simplified, the cost is reduced, the resolution ratio of a detection result is high, and quickness and accuracy are realized.
Owner:SHENZHEN UNI MEDICA TECH
Method for rapid and accurate selective breeding of three-line rice sterile lines by rice genomics technology
ActiveCN108157165AGuaranteed stabilityShortened backcross generationPlant genotype modificationGenomic sequencingAgricultural science
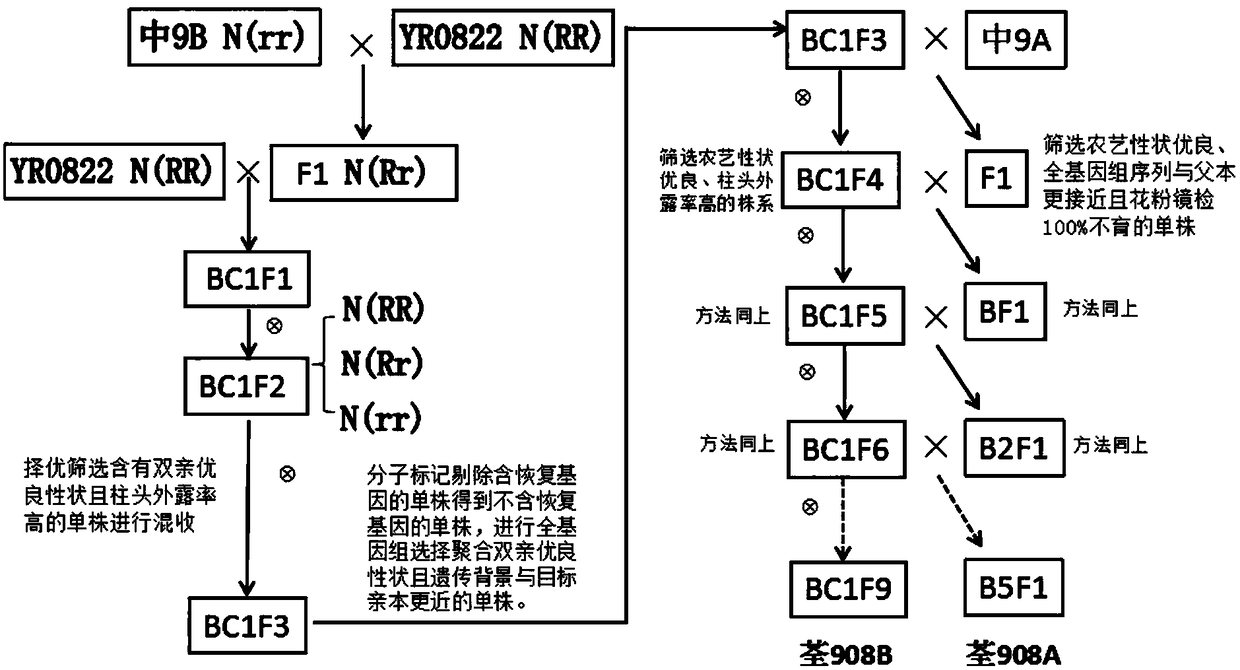
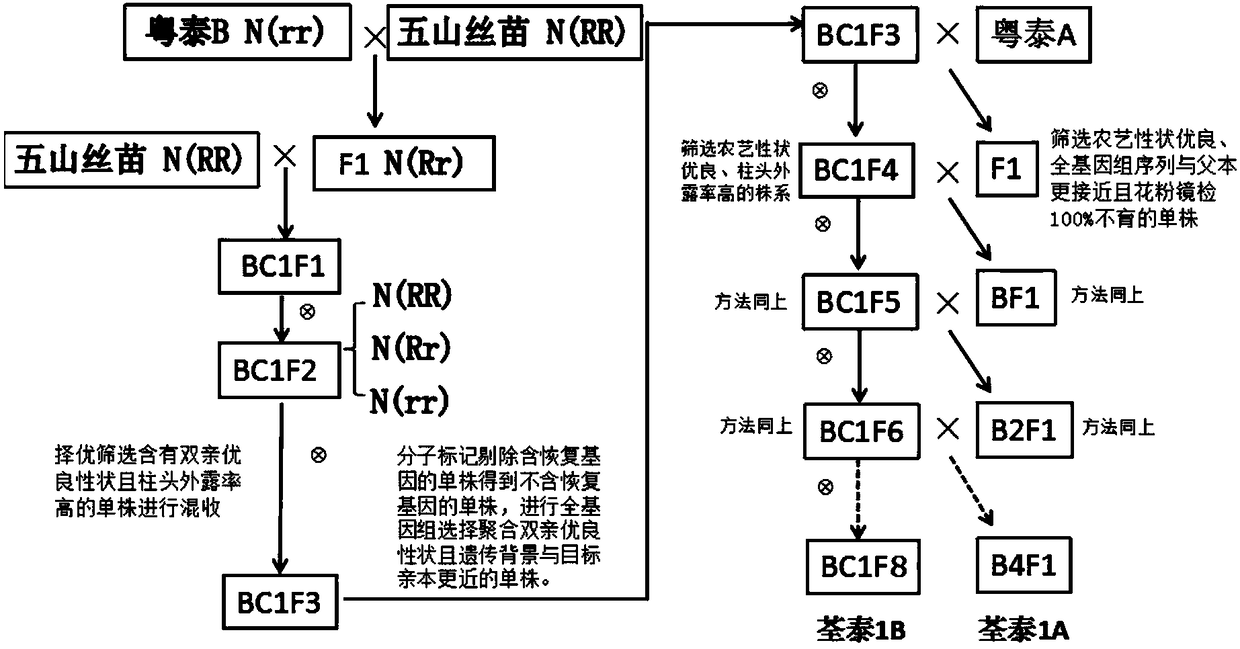
The invention discloses a method for rapid and accurate selective breeding of three-line rice sterile lines by a rice genomics technology. By assisted screening combination of conventional breeding with molecular markers, the method can effectively eliminate fertility restoring genes, and genetic background screening is performed by a genotyping technique based on rice whole genome sequencing, andsterile lines and corresponding maintainer lines having genetic background closer to target parents can be screened. Compared with the prior art, the method greatly shortens improved generations of the maintainer lines and backcross generations of the sterile lines, and the three-line rice sterile lines with excellent comprehensive characters can be accurately selectively bred within only 4-5 years.
Owner:CAS CENT FOR EXCELLENCE IN MOLECULAR PLANT SCI +3
Skin anti-aging ability gene detection primer combination and kit and applications of skin anti-aging ability gene detection primer combination and kit
ActiveCN111455035AAnti agingHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationMass Spectrometry-Mass SpectrometryGenotyping Techniques
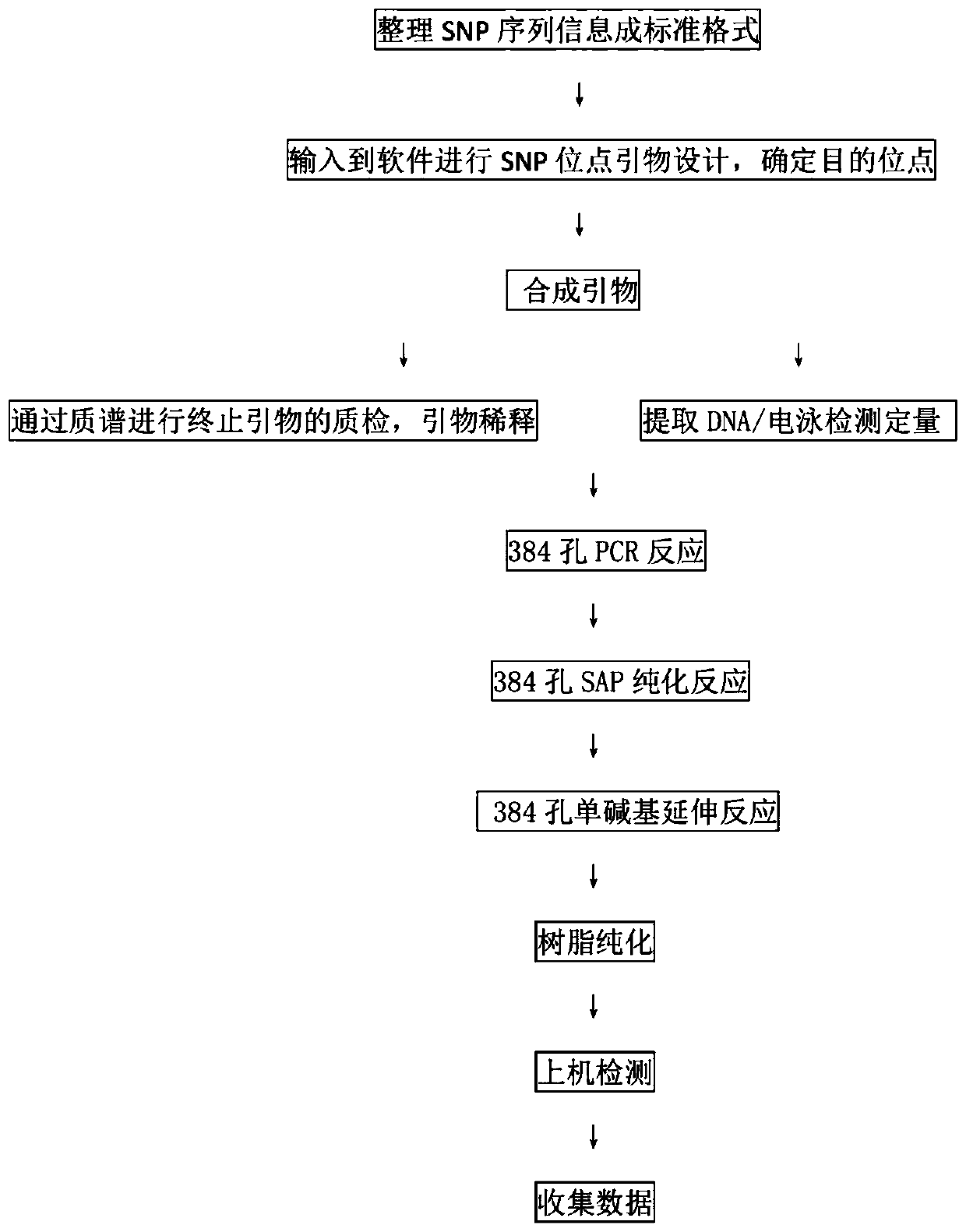
The present invention relates to the technical field of genotyping and particularly to a skin anti-aging ability gene detection primer combination and a kit and applications of the skin anti-aging ability gene detection primer combination and kit. A pair of multiple PCR amplification primers and a single base extension primer are separately designed for each SNP site. All sites are designed into one reaction and can be detected simultaneously. The multiple PCR amplification primer set and the single base extension primer set are used for MassARRAY nucleic acid mass spectrometry analysis systemdetection. The system can be used for rapid analysis of nucleic acid samples with high sensitivity and high accuracy, and a detection accuracy rate is larger than or equal to 99.9%. An SNP typing method for efficiently detecting skin anti-aging ability genes is established, can complete molecular biology genotyping of multiple SNP sites at the same time, has good detection sensitivity, high accuracy, low cost and high practicality, can be used to detect genetic factors that affect skin anti-aging ability, comprehensively evaluates the skin anti-aging ability, and helps promote development ofindividualized services in the feature-beautifying industry.
Owner:广州市普森生物科技有限公司
A method for rapid and precise breeding of three-line rice sterile lines using rice genomics technology
ActiveCN108157165BGuaranteed stabilityShortened backcross generationPlant genotype modificationBiotechnologyGenomic sequencing
The invention discloses a method for quickly and accurately selecting a three-line rice sterile line by using rice genomics technology. The method combines conventional breeding with molecular marker-assisted screening to effectively eliminate fertility restoration genes, and at the same time utilizes the method based on the whole genome of rice Genetic background screening by sequencing genotyping technology can screen and obtain sterile lines and corresponding maintainers whose genetic background is closer to the target parent. Compared with the existing technology, this method greatly shortens the improved generation of the maintainer line and the backcross generation of the sterile line, and can accurately breed a three-line rice sterile line with excellent comprehensive characteristics in only 4-5 years.
Owner:CAS CENT FOR EXCELLENCE IN MOLECULAR PLANT SCI +3
Barley 40K SNP (Single Nucleotide Polymorphism) liquid phase chip
PendingCN114807410AImprove breeding efficiencyNucleotide librariesMicrobiological testing/measurementBiotechnologyHigh throughput genotyping
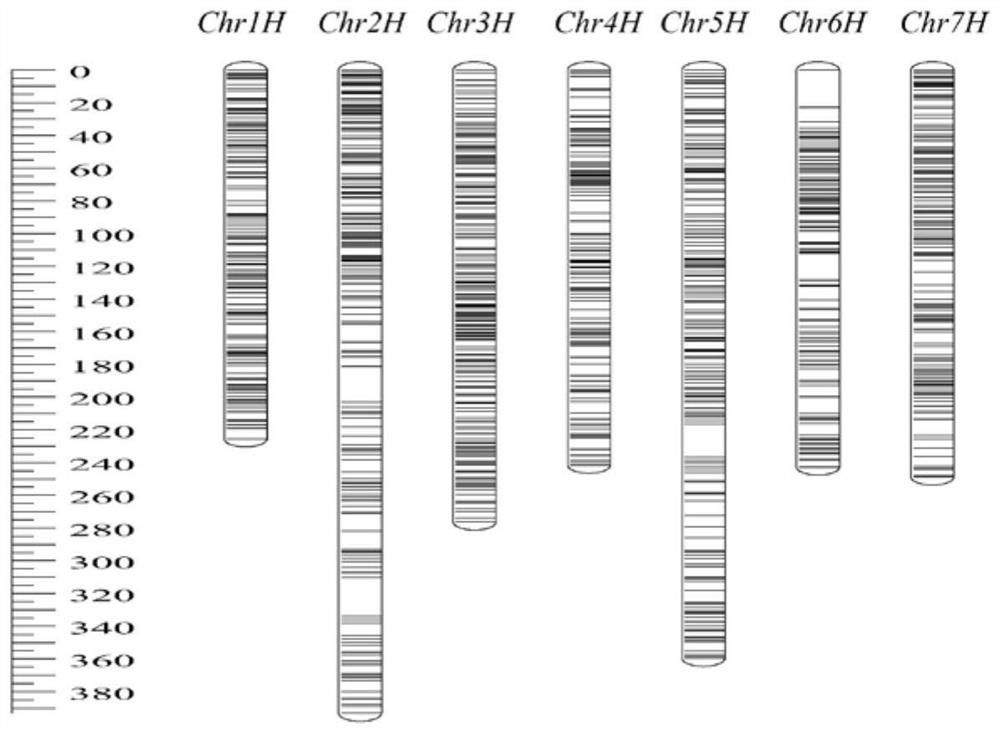
The invention belongs to the technical field of plant molecular markers, and relates to a barley 40K SNP (Single Nucleotide Polymorphism) liquid chip. The barley 40K SNP liquid phase chip is obtained by the following method: 1, selecting barley and highland barley as germplasm materials for re-sequencing, then performing quality control on a sequencing result, mapping with a reference genome, and identifying and screening out SNP loci; 2, according to the indexes of the SNP sites, combining the positions of the sites on chromosomes, analyzing upstream and downstream sequences of the sites, designing sequencing primers, and selecting the SNP sites which can be used for chip development; and 3, developing the barley 40K SNP liquid phase chip by using a targeted sequencing genotyping technology. The barley 40K SNP liquid phase chip belongs to an SNP chip for DNA detection, is a barley whole genome liquid phase chip which is developed based on a targeted capture high-throughput sequencing technology and contains 40519 SNP marker sites, and provides an important tool for barley high-density genetic map construction, barley germplasm identification, high-throughput genotyping, QTL positioning, whole genome association analysis and the like.
Owner:NORTHWEST A & F UNIV
Method for selecting and breeding three-line rice restoring line by rice genomic technology rapidly and accurately
ActiveCN108124764AExpanded genetic backgroundShortening the improved generationPlant genotype modificationGenomic sequencingAgricultural science
The invention discloses a method for selecting and breeding a three-line rice restoring line by a rice genomic technology rapidly and accurately. According to the method, the conventional breeding iscombined with molecular marker auxiliary screening, genetic background screening is conducted by a genotyping technology based on rice whole genome sequencing, and the restoring line which contains ahomozygous sterility restoring gene and has the genetic background closer to a target parent can be screened and obtained. Compared with the prior art, the method has the following advantages: the improved generation of the restoring line is greatly shortened, and the three-line rice restoring line with excellent comprehensive characters can be selected and bred accurately for only 3 to 4 years.
Owner:WIN ALL HI TECH SEED CO LTD +3
Microsatellite primers of red panda gene
InactiveCN106755333AConvenient researchLarge amount of informationMicrobiological testing/measurementDNA/RNA fragmentationMetapopulationGenetic typing
The invention discloses a set of microsatellite primers of red panda gene. The 25 pairs of primers are designed aiming at repeated microsatellite sites from the third base to the sixth base. The phenomenon of slipping chain in primer amplification can be well avoided. The stability is high. The primers have good polymorphism. Through the 25 pairs of red panda microsatellite primers, red panda microsatellite sites with a high content of polymorphism are obtained, a genetic typing technology corresponding to the markers is established, and the primers can be applied to the paternity tests and individual identification of red panda and assist the management of captive populations of red panda.
Owner:CHENGDU RES BASE OF GIANT PANDA BREEDING
Skin whitening capability gene detection primer combination and application thereof
PendingCN111321213AAvoid dullnessReduce dullnessMicrobiological testing/measurementDNA/RNA fragmentationMultiplexHuman DNA sequencing
The invention relates to the technical field of genotyping, in particular to a skin whitening capability gene detection primer combination and an application thereof. The skin whitening capability gene detection primer combination comprises multiplex PCR amplification primer combinations and a single base extension primer combination for detecting 12 SNP sites of 10 skin whitening capability genesof a human genome. Each SNP site is designed with a pair of multiplex PCR amplification primers and a single base extension primer, and all of sites are designed in a reaction to be detected at the same time. According to the SNP typing method for efficiently detecting the skin whitening capability gene, molecular biology genotyping of multiple SNP sites can be completed at the same time, the detection sensitivity is high, the accuracy is high, the cost is low, the practicability is high, the genetic factors influencing skin whitening ability are detected, the comprehensive evaluation of skinwhitening ability is carried out, and the development of individualized services in the beauty industry is promoted.
Owner:广州市普森生物科技有限公司
Gene detection primer combination for acne resistance of skin and application of gene detection primer combination
ActiveCN111321212AGood conditionHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationMultiplexHuman DNA sequencing
The invention relates to the technical field of genotyping, and discloses a gene detection primer combination for acne resistance of a skin and an application of the gene detection primer combination.The gene detection primer combination comprises a multiplex PCR amplification primer group and a single base extension primer group which are used for detecting eight SNP loci of five acne resistancegenes of a human genome. Each SNP locus is designed with a pair of multiplex PCR amplification primers and a single base extension primer, and all the sites are designed into a reaction and can be detected simultaneously. According to the invention, an SNP typing method for efficiently detecting the anti-acne gene of the skin is established, and molecular biological genotyping of a plurality of SNP loci can be completed at the same time. The detection sensitivity is good, accuracy is high, cost is low, and practicability is high. The SNP typing method can be used for detecting genetic factorsinfluencing acne resistance of the skin and comprehensively evaluating acne resistance of the skin, and promotes development of individual service in the beauty industry.
Owner:广州市普森生物科技有限公司
Microsatellite primer combination for identifying genetic relationship between eggs of Lophophorus lhuysii in same bird house and application
ActiveCN112980971AHigh polymorphic contentImprove stabilityMicrobiological testing/measurementAgainst vector-borne diseasesBiotechnologyEggshell
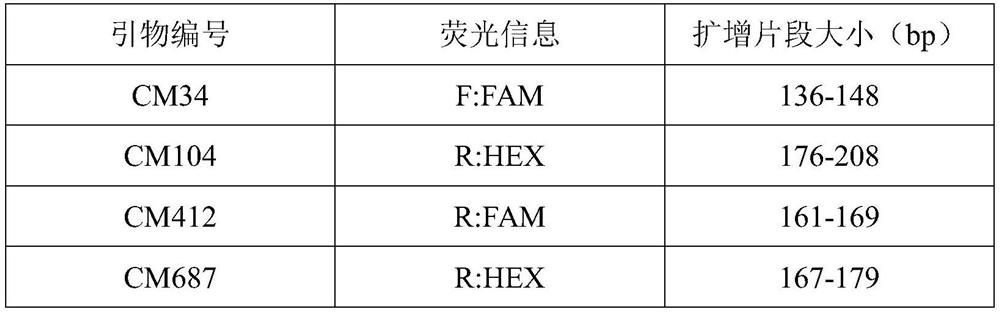
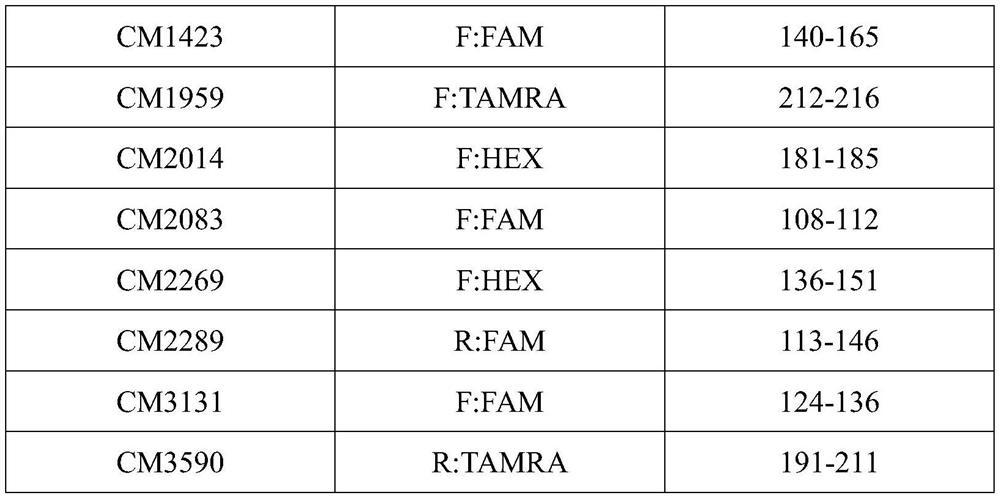
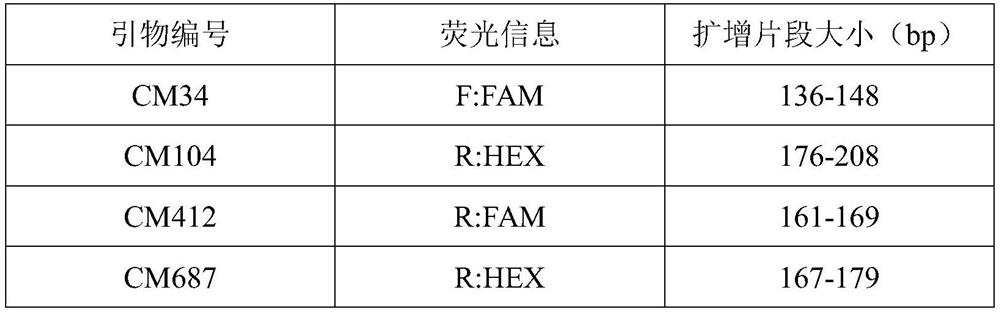
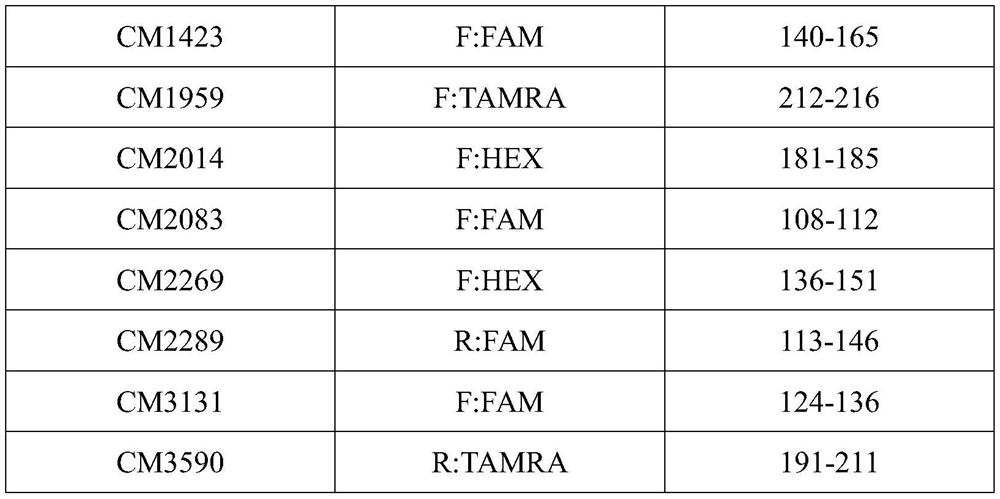
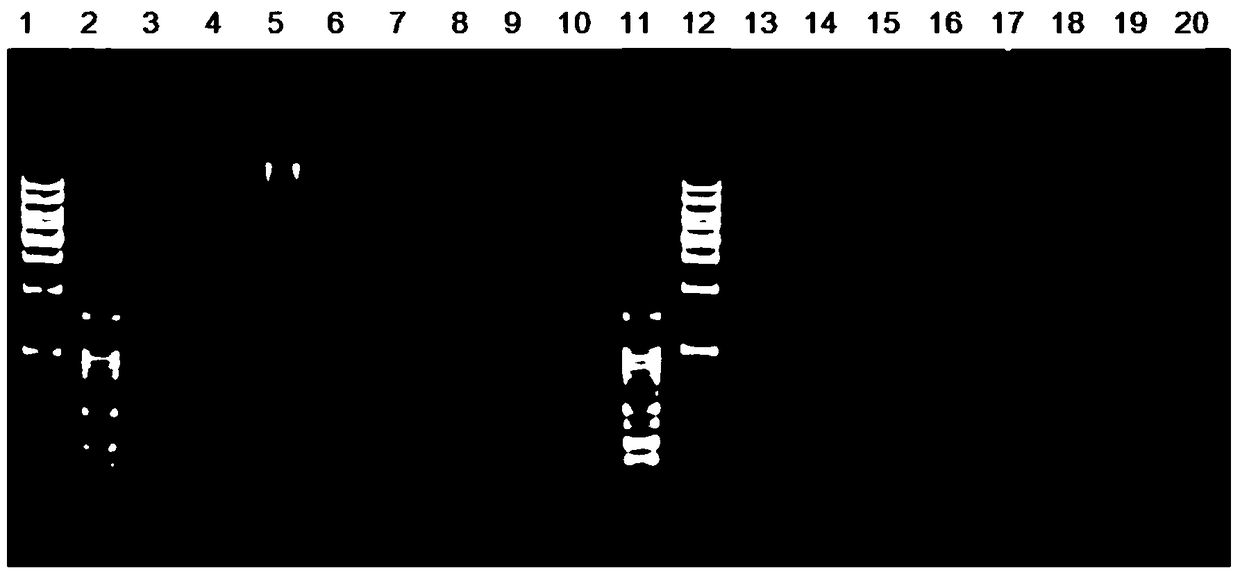
The invention discloses a microsatellite primer combination for identifying the genetic relationship between eggs of Lophophorus lhuysii in the same bird house and application. The primers comprise CM34, CM104, CM412, CM687, CM1423, CM1959, CM2014, CM2083, CM2269, CM2289, CM3131 and CM3590, and the nucleotide sequences of the primers are sequentially shown as SEQ ID NO.1-24. The primers can be stably amplified in the eggshell DNA of Lophophorus lhuysii, the microsatellite sequence of Lophophorus lhuysii with high polymorphism content and good repeatability is obtained through the 12 pairs of primers, the genotyping technology corresponding to the markers is established, and identification of three genetic relationship conditions of sib, half-sib or non-paternity between the eggs of Lophophorus lhuysii in the same bird house can be realized.
Owner:CHENGDU RES BASE OF GIANT PANDA BREEDING
Multi-SNP locus genetic typing method based on nMALDI-TOF technology
PendingCN114525326AShorten experiment timeRealize multi-gene locus detectionMicrobiological testing/measurementEnzyme digestionHybridization reaction
The invention belongs to the technical field of SNP (Single Nucleotide Polymorphism) locus genotyping, and particularly relates to a multi-SNP locus genotyping method based on an nMALDI-TOF (Independent Matrix Assisted Laser Desorption / Ionization Time of Flight) Comprising the following steps: designing and synthesizing a probe and a PCR (Polymerase Chain Reaction) amplification primer aiming at a plurality of SNPs of DNA of a sample to be detected; hybridizing the probe and the sample DNA; adding extension and connection reaction liquid into a hybridization reaction product, uniformly mixing, and carrying out extension and connection; adding excision enzyme mixed liquor into extension and connection reaction products, uniformly mixing, and carrying out enzyme digestion; adding S5amp into an enzyme digestion reaction product; uniformly mixing the mixed solution of the N7 primer, purified water and the PCR premixed solution, and carrying out PCR reaction; and after desalting a PCR reaction product, loading a sample to MALDI-TOF-MS for detection to obtain a detection result. On the basis that the result is accurate, the operation time can be shortened by half.
Owner:THE SECOND PEOPLES HOSPITAL OF SHENZHEN
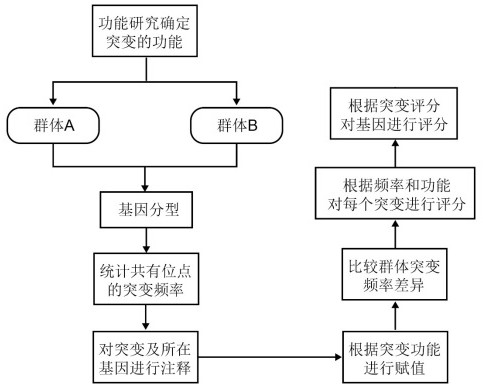
Population genetic difference comparison method based on mutation function
The invention discloses a method for comparing the genetic variation difference aiming at the important function mutation of the population, which comprises the steps of evaluating a mutation functionby using prediction software or an actual function verification test to obtain a set of function mutations and then giving higher weight values to the function mutations; and calculating the gene frequency of a polymorphic site of certain population after genotype information of each individual in the population is obtained by utilizing a second-generation sequencing technology or a genotyping technology such as an SNP chip, so that the difference degree of genetic polymorphism taking a gene as a unit among races can be calculated in batches by utilizing a calculation formula of the method. According to the method, a genetic unit rises from a single polymorphic site to a single gene, so that the difference degrees of all functional mutations of a certain gene among different populations can be compared to predict the difference of some gene-related phenotypes among different populations and guide the development of gene-related genetic research among different populations.
Owner:XIANGYA HOSPITAL CENT SOUTH UNIV
A set of microsatellite primers for red panda genes
InactiveCN106755333BAvoid slipping chainsEasy to manageMicrobiological testing/measurementDNA/RNA fragmentationGenetic typingGene
Owner:CHENGDU RES BASE OF GIANT PANDA BREEDING
Mutation site associated with total litter size character of sow and application of mutation site
ActiveCN114875028AAccelerate localization selection and breeding goalsAccelerate cultivationFood processingMicrobiological testing/measurementNucleotidePig breeding
The invention discloses a pig miR-23a gene core promoter sequence and a mutation site obviously associated with the total litter size character of a sow, the site is located at the 261 basic group position of the pig miR-23a gene core promoter (namely the 65307715 nucleotide position of a second chromosome reference genome), and the site is Cgt; the total litter size of the TT type multiparous sow is higher than that of CC type and CT type sows. The total litter size character is a low-heritability breeding character, the genetic progress is slow by adopting a conventional breeding technology, and as the mutation site is obviously associated with the total litter size character, the high-yield sows can be selected and reserved by detecting the site by adopting a simple and convenient genetic typing technology; the method has important values for accelerating the breeding of high-yield female parent new strains, early completing the localized breeding goal of large white pigs, improving the core competitiveness of the live pig breeding industry in China and the like.
Owner:NANJING AGRICULTURAL UNIVERSITY
Analysis method of porcine SNP marker loci based on sequencing genotyping technology
ActiveCN106566872BGuaranteed accuracyLow parting costMicrobiological testing/measurementGenomic sequencingEnzyme digestion
A pig SNP marker site analyzing method based on a sequence-based typing technique is provided. The method includes steps of (1) predicting a distribution situation of digested fragments obtained by subjecting a pig genome to double-enzyme digestion using EcoRI and MspI, (2) designing general linkers, bar code linkers and PCR amplification primers according to the digestion recognition sequences of the EcoRI and MspI, (3) constructing a simplified genome sequencing library, (4)performing computer sequencing by utilizing the library constructed in the step (3), and (5) acquiring SNP marker sites according to the sequencing result. The digestion composition used in the method is suitable for all pig varieties. The composition is utilized to simplify genome sequencing, and a number of SNPs acquired can be higher than a number of SNPs on a chip available in the market at present. The method can be used for whole-genome correlation analysis and research and genome selection research, and the typing cost of a single body can be lower than that of a chip popular at present.
Owner:CHINA AGRI UNIV
A gene mutation site for weeding out low fecundity Hu sheep and its application
ActiveCN109207602BReduced reproductive capacityMicrobiological testing/measurementDNA/RNA fragmentationGenes mutationAnimal science
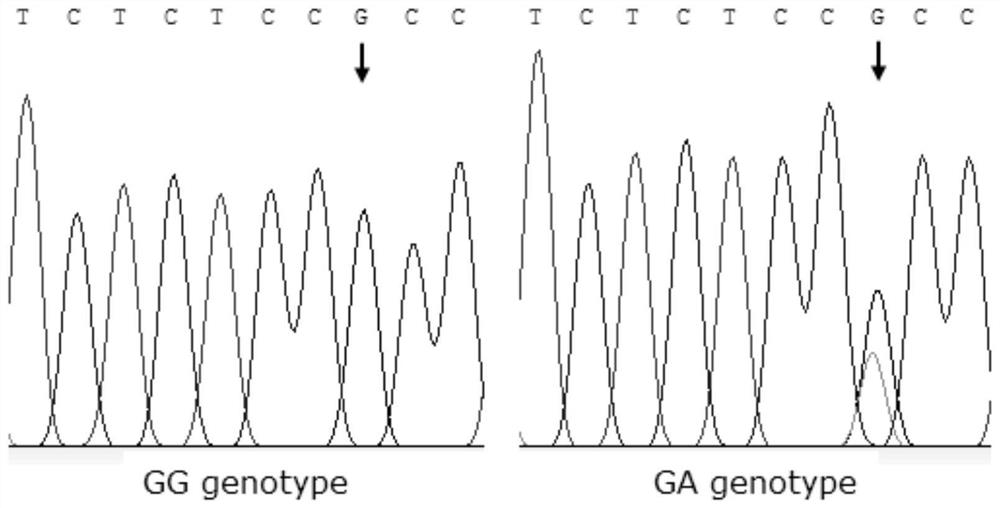
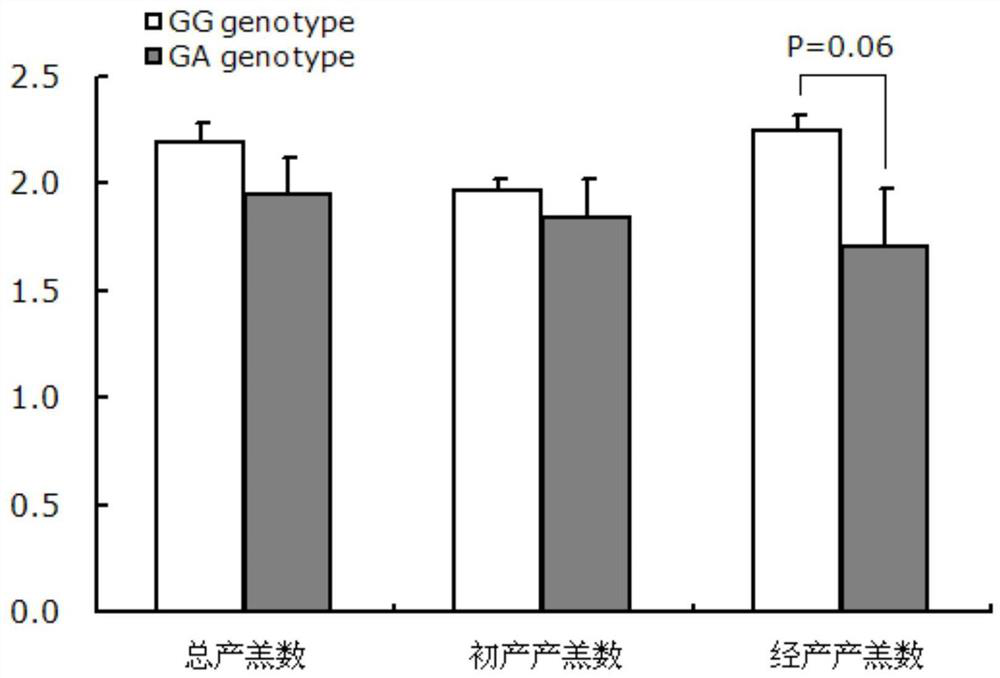
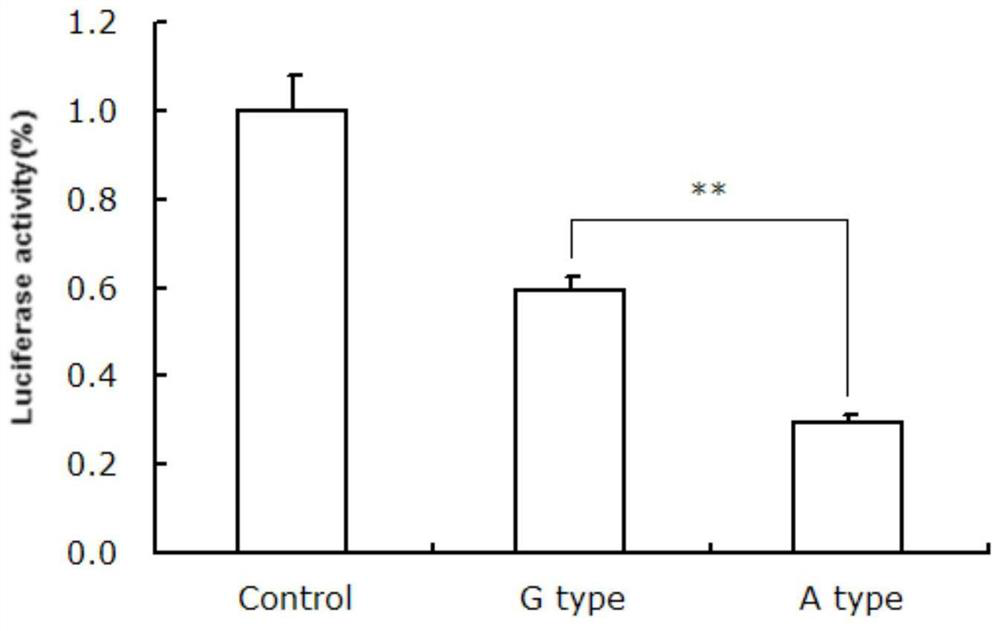
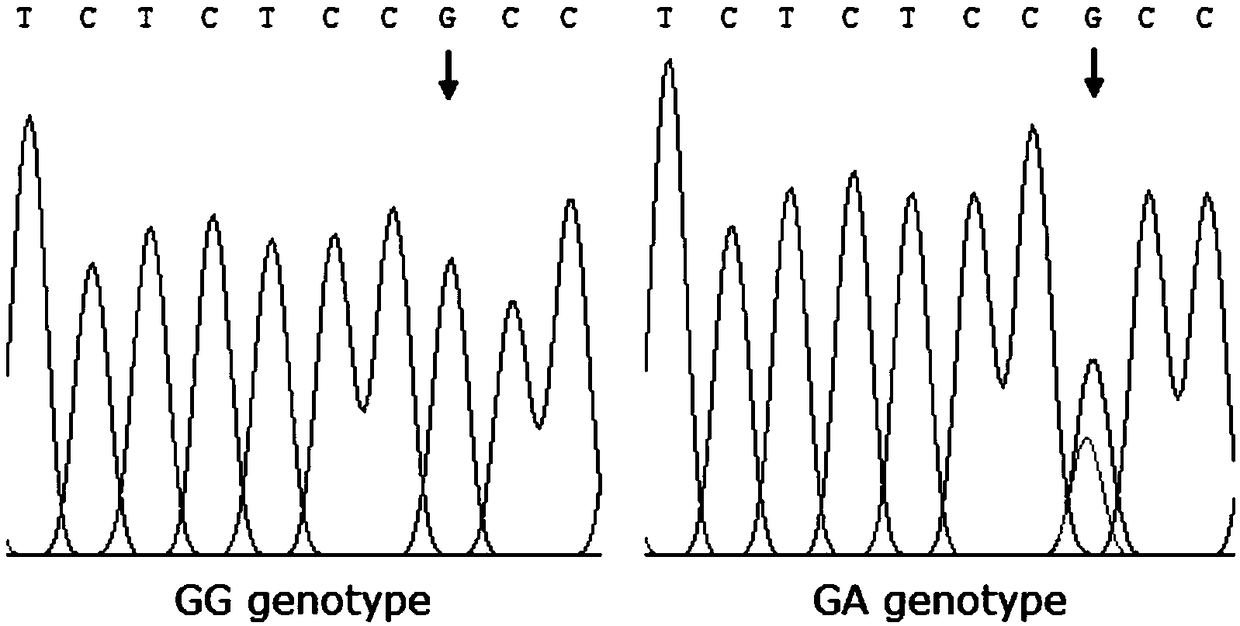
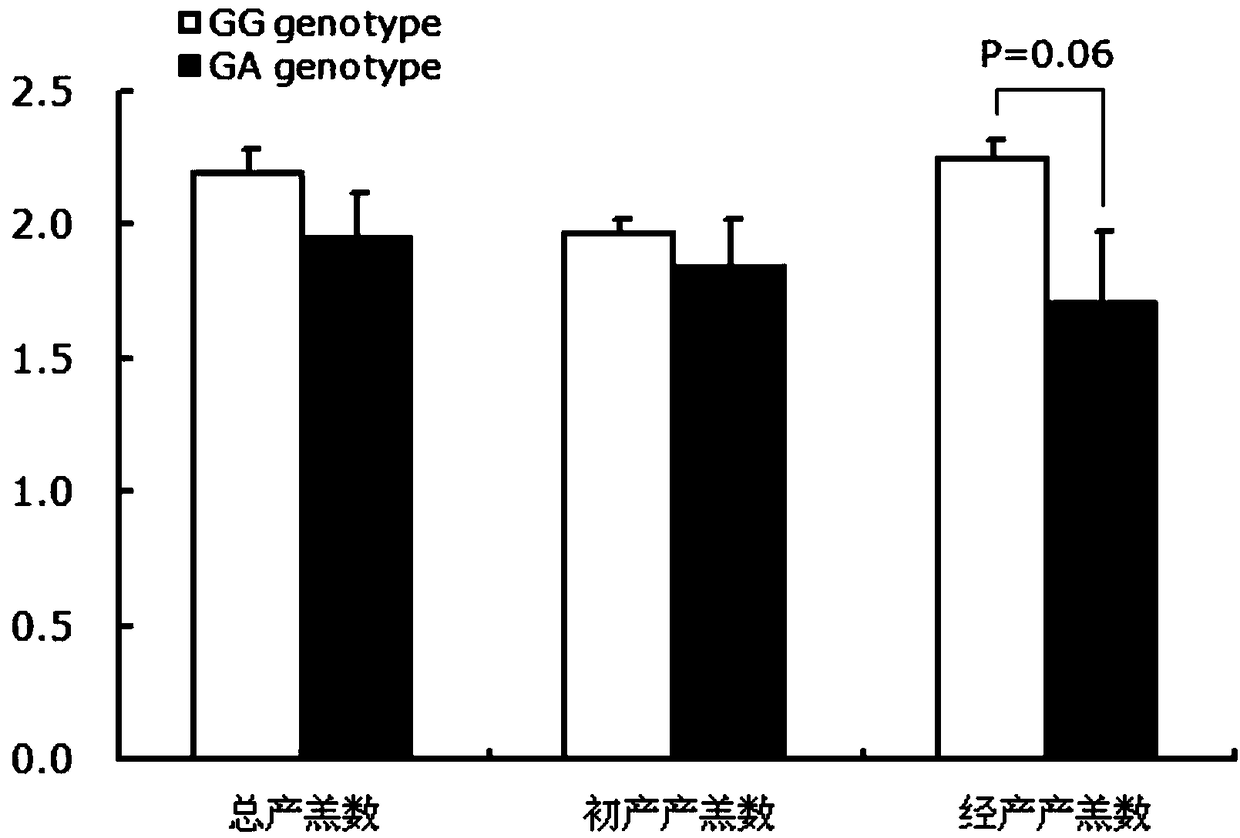
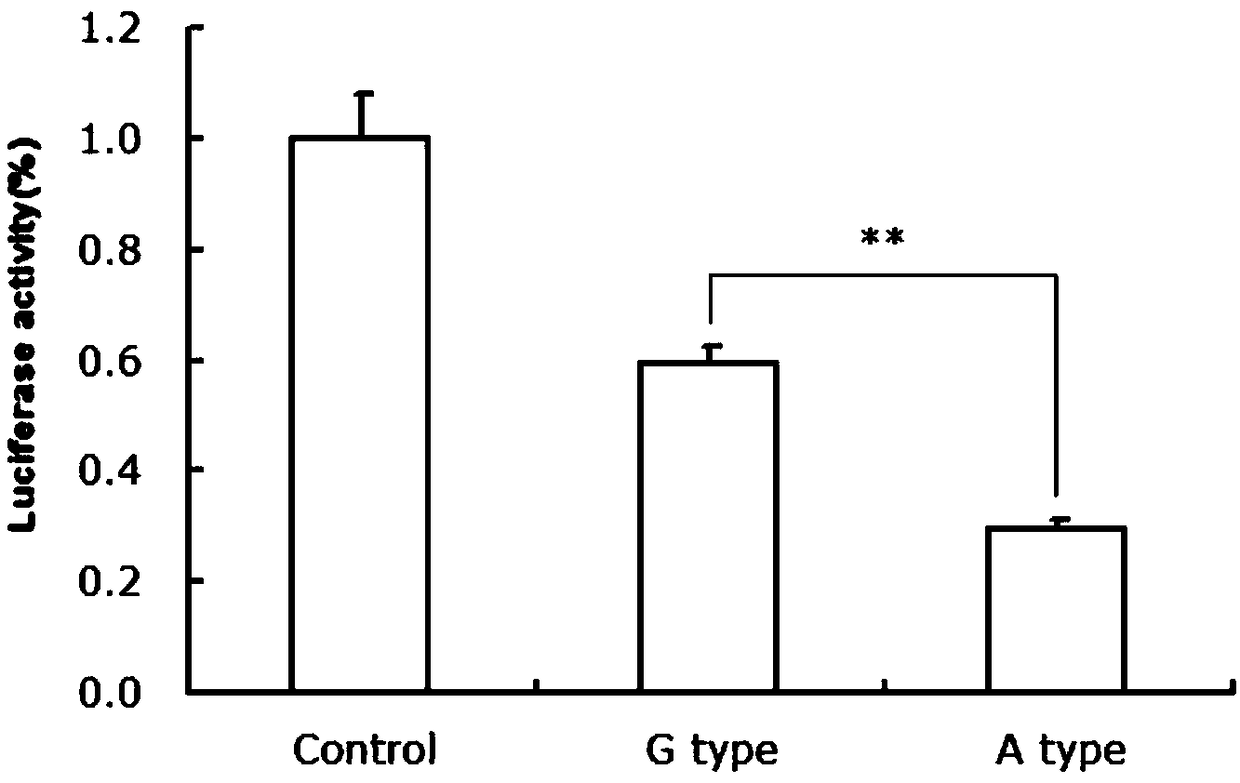
The invention discloses a gene mutation site for eliminating low fecundity Hu sheep and its application. The mutation site is located at the 1354th nucleotide of the 3'-UTR of Hu sheep BMPR1B gene, and the site is G>A The average number of lambs per litter in GA-type multiparous Hu sheep ewes was 0.54 lower than that of GG-type individuals, and the transcriptional activity of BMPR1B gene 3'‑UTR of A-type was only half that of G-type. The reproductive trait of livestock is a low heritability trait, and the genetic progress of conventional breeding technology is relatively slow. Because the mutation site causes the reproductive performance of multi-bred Hu sheep ewes to be significantly reduced, simple genotyping technology is used to detect this site. The individuals with low fertility genotypes can be eliminated, and the individuals with high fertility genotypes can be selected, which has important economic value in accelerating the progress of breeding of high fertility Hu sheep populations or strains.
Owner:NANJING AGRICULTURAL UNIVERSITY
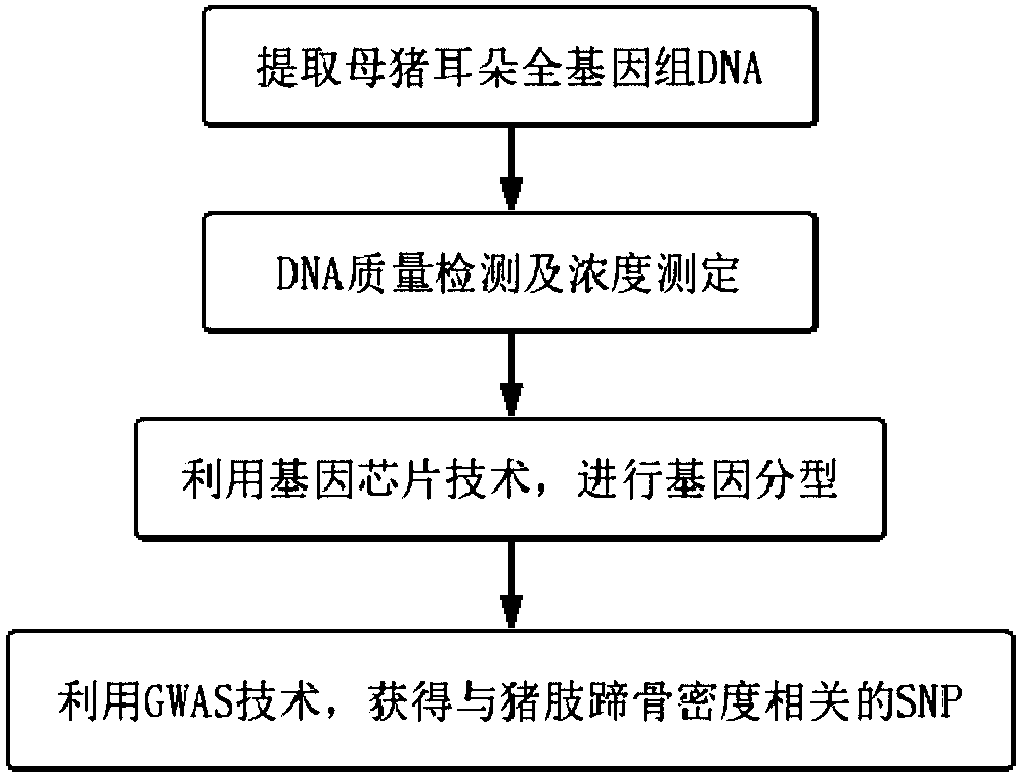
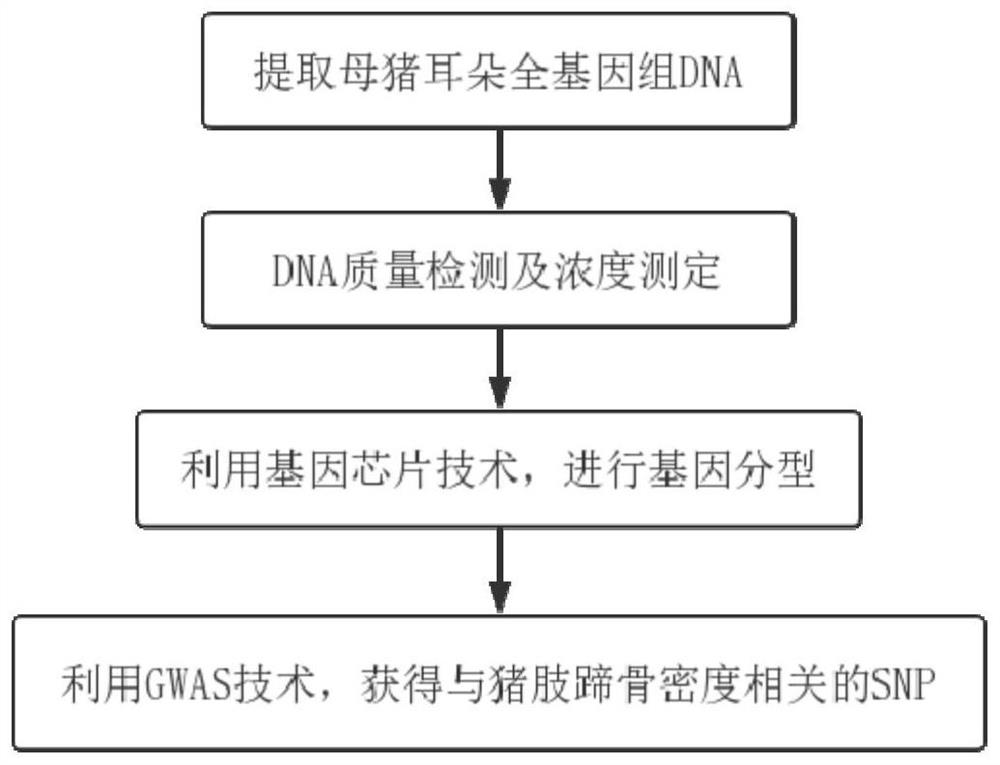
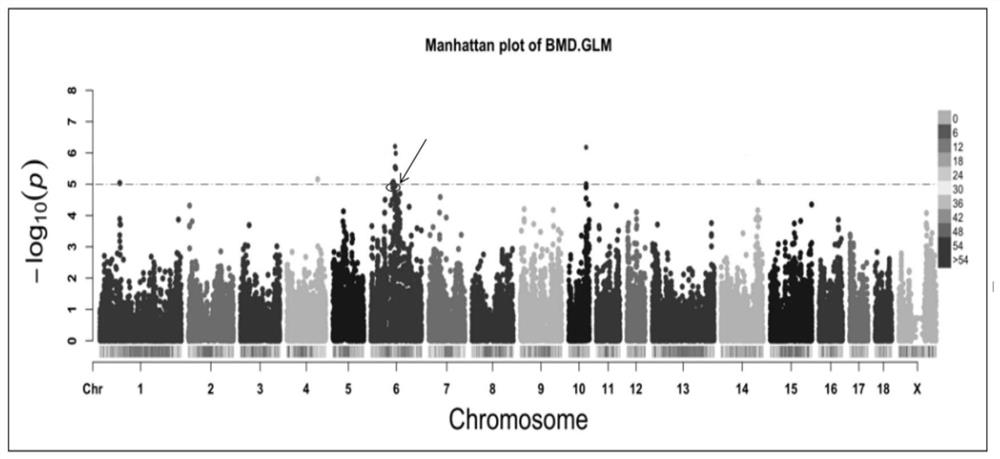
SNP molecular marker of FAM131C gene associated with bone density of limb and hoof of sow
ActiveCN108384864AHigh sensitivityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationEnsembl databaseBone density
The invention belongs to the field of animal molecular marker screening, and particularly relates to an SNP molecular marker of an FAM131C gene associated with bone density of a limb and hoof of a sow. An FAM131C gene segment of a gene upstream and downstream 100bp sequence with an accession number of ASGA0099279 is obtained by a genotyping technology and referring to an Ensembl database, and thesequence is as shown in SEQ ID NO:1. By conducting typing on the FAM131C gene using a gene chip technology, associated markers of the limb and hoof of the sow are obtained by screening, the sequence is as shown in SEQ ID NO:1, and the R of the 101 basic group of the sequence is C or T, which leads to the polymorphism of a nucleotide sequence. The gene can be taken as the molecular marker usedfor the detection associated to the sow bone density, and when the 101 nucleotide on SEQ ID NO:1 is C, the limb and hoof of the sow have higher bone density. The invention provides a new SNP marker for marking assisting selection of sow bone density.
Owner:HUAZHONG AGRI UNIV
Complete sequence of porcine BMP7 gene 3'-UTR, mutation sites associated with reproductive traits and application of mutation sites
PendingCN112301135ASpeed up breeding progressMicrobiological testing/measurementDNA/RNA fragmentationMedicinePig reproduction
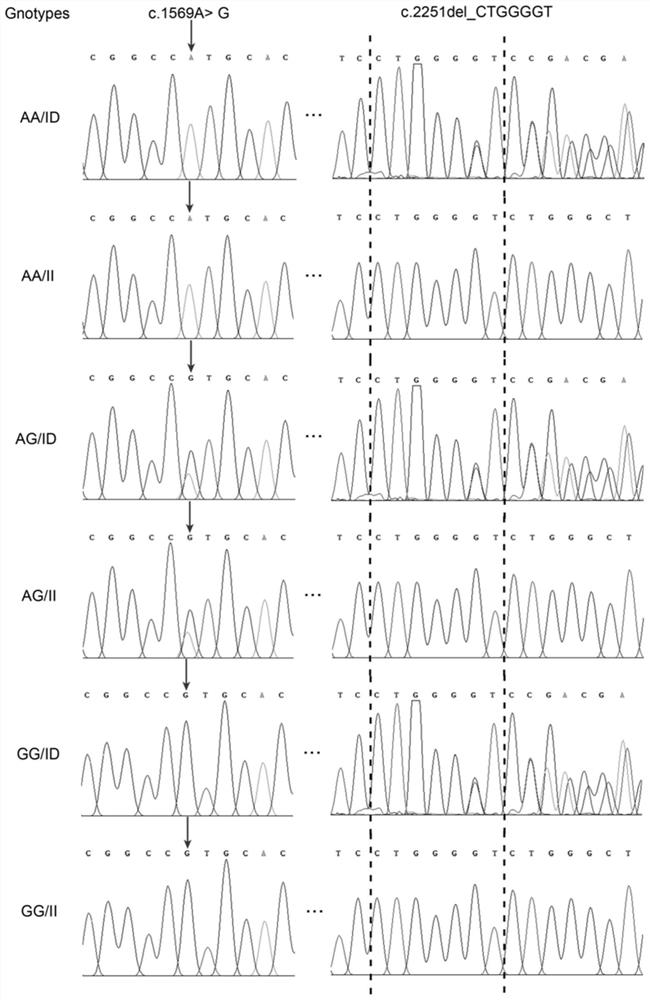
The invention discloses a complete sequence of a porcine BMP7 gene 3'-UTR, mutation sites associated with reproductive traits and application of the mutation sites. The two mutation sites significantly associated with the porcine reproductive traits are located at the 273rd site and the 955th site of the porcine BMP7 gene 3'-UTR respectively, wherein the first site is A>G mutation named as c.1569A>G, and the second mutation is insertion / deletion mutation named as c.2251indelCTGGGGT. The sow reproductive traits are low-heredity traits; the genetic progress is relatively slow due to the adoptionof a conventional breeding technology; due to the fact that the combined genotype of the mutation sites is remarkably associated with the total litter size of a sow and is remarkably close to the live litter size, the two sites are detected by adopting a simple and convenient genotyping technology, so that high-yield genotype individuals can be selected and remained, and low-yield genotype individuals are eliminated; and therefore the mutation sites have important economic value for selecting and remaining the high-yield individuals and improving the reproductive performance of sow groups.
Owner:NANJING AGRICULTURAL UNIVERSITY
SNP molecular marker of fam131c gene related to bone mineral density of sow limbs and hoofs
ActiveCN108384864BHigh sensitivityStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationAnimal scienceBone density
The invention belongs to the field of animal molecular marker screening, in particular to the SNP molecular marker of the FAM131C gene related to the bone density of sow limbs and hooves. Through genotyping technology and referring to the Ensembl database, the FAM131C gene fragment with the accession number ASGA0099279 gene upstream and downstream 100 bp sequence was obtained, and the sequence is shown in SEQ ID NO:1. The FAM131C gene was typed using gene chip technology, and the markers related to the bone density of sow limbs and hooves were screened. Its sequence is shown in SEQ ID NO: 1, and the R at the 101st base of the above sequence is C or T , leading to polymorphisms in nucleotide sequences. It can be used as a molecular marker for detection of porcine bone density. When the 101st nucleotide on SEQ ID NO: 1 is C, the sow's limbs and hooves have higher bone density. The invention provides a new SNP marker for marker-assisted selection of porcine bone density.
Owner:HUAZHONG AGRI UNIV
Primer combination for detecting genes related to skin moisturizing abilities and application of primer combination
PendingCN111378762AReduce drynessHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationGenomeBioinformatics
The invention relates to the technical field of genotyping, in particular to a primer combination for detecting genes related to skin moisturizing abilities and application of the primer combination.The primer combination includes a multiple-PCR-amplification primer group and a single base extension primer group which are used for detecting 12 SNP sites of 7 skin moisturizing ability genes in thehuman genome, wherein a pair of multiple-PCR-amplification primers and a single base extension primer are designed for each SNP site, and all the sites are designed into one reaction, and can be detected simultaneously. An SNP typing method for efficiently detecting the genes relates to skin moisturizing abilities is established, molecular biology genotyping of multiple SNP sites can be completedat the same time, and the method has good detection sensitivity, high accuracy, low cost and high practicability, can be applied to detection of genetic factors which influence skin moisturizing abilities, and applied to comprehensive evaluation of skin moisturizing abilities, so that development of personalized services in the beauty industry is promoted.
Owner:广州市普森生物科技有限公司
Primer Combination and Its Application for Detection of Skin Anti-Acne Ability Gene
ActiveCN111321212BGood conditionHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationHuman DNA sequencingMedicine
Owner:广州市普森生物科技有限公司
A gene mutation site for eliminating low-fertility Hu sheep and application thereof
ActiveCN109207602AReduced reproductive capacityMicrobiological testing/measurementDNA/RNA fragmentationLow fertilityGenotype
The invention discloses a gene mutation site for eliminating low-fertility Hu sheep and application thereof, wherein the mutation site is located at 1354 site of the 3'-UTR of the Hu sheep BMPR1B gene, the mutation is G to A, the average litter size per litter of Hu ewes with GA genotype is 0.54 lower than that of Hu ewes with GG genotype, and the transcriptional activity of 3'-UTR of the BMPR1B gene is only half of that of G-type. Animal reproductive trait is a low heritability trait, genetic progress is slow with conventional breeding techniques, As that mutation site lead to obvious reduction of reproductive performance of the breeding Hu sheep ewe, the low-fecundity genotype individuals can be eliminated and the high-fecundity genotype individuals can be retain by adopting a simple genotyping technique to detect the mutation site, and the mutation site has important economic value for speeding up the breeding progress of the high-fecundity Hu sheep population or strain.
Owner:NANJING AGRICULTURAL UNIVERSITY
Microsatellite Primer Combination and Application for Identification of Genetic Relationship in Eggs of Green-tailed Pheasant's Pheasant
ActiveCN112980971BHigh polymorphic contentImprove stabilityMicrobiological testing/measurementAgainst vector-borne diseasesBiotechnologyEggshell
Owner:CHENGDU RES BASE OF GIANT PANDA BREEDING
Method for Determining Sequencing Digestion Combination in Sequencing Genotyping Technology
ActiveCN105368930BGuaranteed accuracyLow parting costMicrobiological testing/measurementEnzyme digestionRestriction enzyme digestion
The invention provides a determining method for sequencing enzyme digestion combination in a sequencing genotyping technology. The determining method comprises the following steps that 1, restriction enzyme digestion site predicting is performed on a target genome, and the number of enzyme digestion segments obtained through different enzyme digestion modes is counted; 2, a joint sequence and a PCR amplification primer sequence at the two ends of each enzyme digestion segment are designed according to the predicted enzyme digestion segments in the various enzyme digestion modes in the step 1; 3, sequencing libraries are constructed through a GBS technology for the different enzyme digestion modes; 4, sequencing is performed through the sequencing libraries constructed in the step 3; 5, SNP marker sites are obtained according to sequencing results; 6, the specific enzyme digestion combination for the target genome is determined according to the number of the SNP marker sites and the enzyme digestion segment sizes which are obtained through different enzyme digestion combinations.
Owner:CHINA AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com