Barley 40K SNP (Single Nucleotide Polymorphism) liquid phase chip
A liquid phase chip, barley technology, applied in the fields of instruments, nucleotide libraries, library creation, etc., can solve the problems of high cost and high technical threshold, and achieve the effect of improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
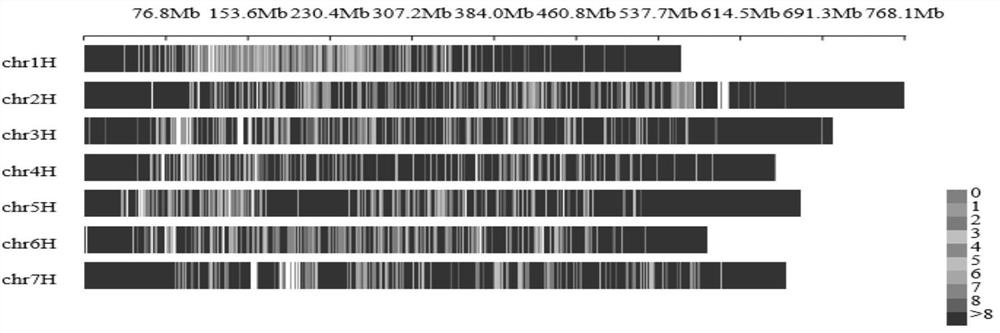
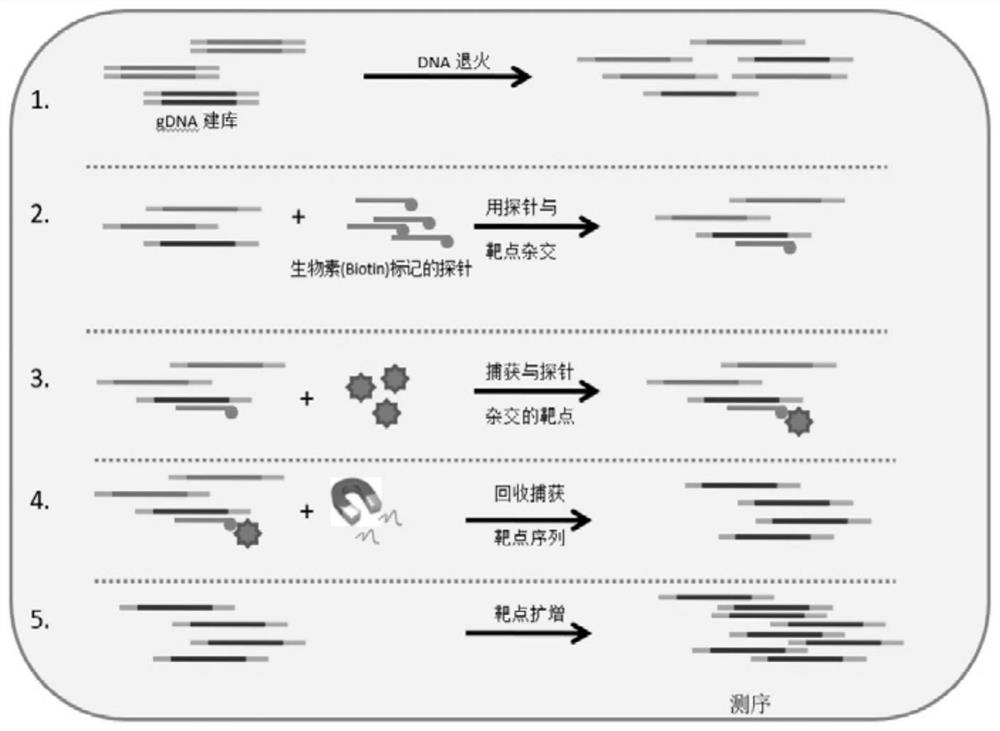
[0037] A barley 40K SNP liquid phase chip is obtained through the following main processes: based on whole-genome resequencing of barley materials from different ecological regions and different types of the world, more than 45 million SNP loci have been obtained; The index, the position of the binding site on the chromosome, analyze the upstream and downstream sequences, and select 40,519 SNP sites with strong representation, good polymorphism, and uniform chromosomal distribution that can be used for chip development; finally, targeted sequencing genes are used. Genotyping by Targeted Sequencing (GBTS) technology, which is a liquid-phase chip technology that only performs targeted deep resequencing of these 40,519 target sites, developed a 40K liquid-phase SNP chip of barley, and named the chip For the Barley SNP 40K Array.
Embodiment 2
[0039] A barley 40K SNP liquid phase chip is obtained by the following method.
[0040] (1) Barley resequencing and identification and screening of SNP loci
[0041]S1. In order to ensure the representativeness of barley materials, according to the analysis of agronomic traits combined with molecular markers of nearly 2,000 materials stored in the laboratory, and according to regional sources and types, 155 representative barley germplasms were selected as materials. These materials come from a wide range of sources. , strong representation, including 43 wild barley in different ecoregions worldwide (including South Levant, North Levant, East Levant, Iran, Caucasus, etc.), 54 barley farms (Middle East, Eastern Europe, etc.) , Australia, the United States, China, etc.), 27 cultivars (20 countries such as the United States, Canada, the United Kingdom, Germany, etc.) and 31 highland barley materials (wild, local and cultivated); select 10-15 full and complete trees of each line ...
Embodiment 3
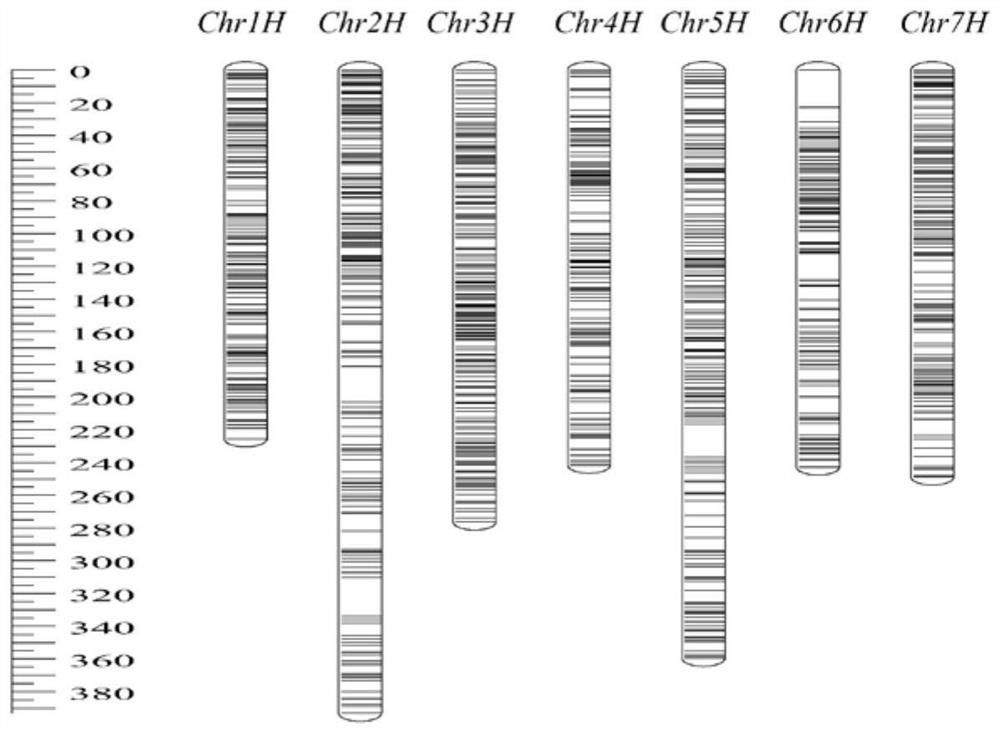
[0061] In order to further evaluate the value of the 40K liquid-phase SNP chip in the actual molecular breeding of barley, it is now applied to the construction of high-density genetic map of barley (such as image 3 shown). The specific process and results are as follows.
[0062] The liquid phase SNP chip of the present invention was used to genotype 125 individuals of the Steptoe×Morex DH population, and a total of 16972 polymorphism sites were obtained, thereby constructing the first high-density SNP genetic map of barley; in the genetic map, The total genetic distance is 2023.33cM, including 7 linkage groups, and the average map distance is 2.10cM (such as image 3 In addition, the number of markers distributed on the 7 chromosomes of barley is also relatively average, and the number of markers on each chromosome ranges from 103 to 165. Among them, chromosome 1 is the shortest at 225.38cM, and chromosome 2 is the longest. Based on this map, QTL mapping was carried out f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com