Method, database and primer pair for identifying herbal medicinal materials
A database and primer technology, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as inability to achieve rapid identification, time-consuming and cost-consuming, and basic mixing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0268] Primer pair optimization for plants:
[0269] According to the sequence between the transcribed 18S ribosomal RNA region and the transcribed 28S ribosomal RNA region of the plant genome DNA sequence, 12 primers were designed, as shown in Table 1.
[0270] Table 1, the primer of the embodiment of the present invention
[0271]
[0272] The combination of the above-mentioned different forward primers and reverse primers can reproduce the sequence covering the part of the transcribed 18S ribosomal RNA region of the plant genome DNA sequence, the sequence of the transcribed spacer 1 (ITS1), and the sequence of the transcribed 5.8S ribosomal RNA region , the sequence of the transcribed spacer 2 (ITS2) and / or the sequence of a part of the transcribed 28S ribosomal RNA region, or a part of the sequence that can be identified between the transcribed 18S ribosomal RNA region and the transcribed 28S ribosomal RNA region.
[0273] After different combinations and optimization ...
Embodiment 2
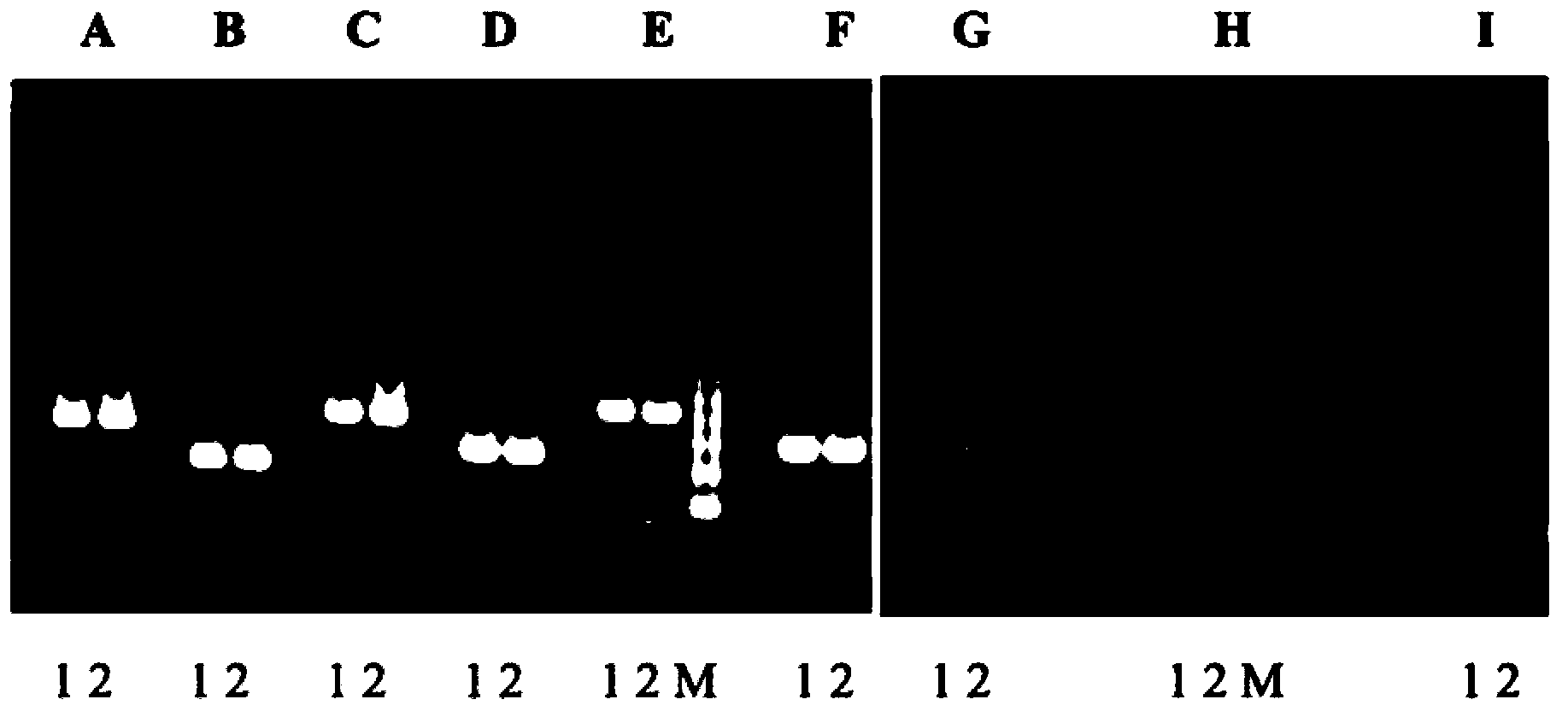
[0286] Take the primer pair ITRI-F2 (SEQ ID: 1) / ITRI-470R (SEQ ID: 10) and the primer pair ITRI-400F (SEQ ID: 4) / ITRI-R2 (SEQ ID: 7) as an example , the above two primer pairs were used to perform polymerase chain reaction on DNA samples of various herbal medicines, and the products were analyzed by electrophoresis to explore their applicability and wide application. Complete DNA fragment replication of at least 95 families, 186 genera and 224 kinds of medicinal materials to provide HRM map database construction and analysis. The results of the primer pair ITRI-F2 (SEQ ID: 1) / ITRI-470R (SEQ ID: 10) and the primer pair ITRI-400F (SEQ ID: 4) / ITRI-R2 (SEQ ID: 7), These are shown in Figures 2a to 2q and Figures 3a to 3q, respectively. The medicinal materials represented by the symbols in Figures 2a to 2q and Figures 3a to 3q are shown in Table 2.
[0287] Table 2, medicinal materials represented by labels in Figures 2a to 2q and Figures 3a to 3q
[0288]
[0289]
[0290]...
Embodiment 3
[0295] High-resolution melting analysis (HRMA) map database construction of herbal medicinal materials:
[0296] The primer pairs of ITRI-F2 / ITRI-470R and ITRI-400F / ITRI-R2 were used to copy the fragments of various herbal medicinal materials, and then HRM analysis was performed to construct a unique ratio of 224 medicinal materials in 95 families, 186 genera (Table 2). to graph databases. Primer pair ITRI-F2 (SEQ ID: 1) / ITRI-470R (SEQ ID: 10) for the high-resolution double-strand segregation dynamics analysis of fragments copied from different plant medicinal materials is selected for each family The methods of plants are representatively shown in Figures 99 to 193; and, the primer pair ITRI-400F (SEQ ID: 4) / ITRI-R2 (SEQ ID: 7) replicated fragments of different plant medicinal materials The high-resolution double-strand segregation kinetic analysis maps are also representatively shown in Figures 99 to 193 in the way of selecting one plant per family.
[0297] In addition, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com